Structural and Comparative Analysis of the Complete Chloroplast Genome of a Mangrove Plant: Scyphiphora Hydrophyllacea Gaertn
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

"National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary."
Intro 1996 National List of Vascular Plant Species That Occur in Wetlands The Fish and Wildlife Service has prepared a National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary (1996 National List). The 1996 National List is a draft revision of the National List of Plant Species That Occur in Wetlands: 1988 National Summary (Reed 1988) (1988 National List). The 1996 National List is provided to encourage additional public review and comments on the draft regional wetland indicator assignments. The 1996 National List reflects a significant amount of new information that has become available since 1988 on the wetland affinity of vascular plants. This new information has resulted from the extensive use of the 1988 National List in the field by individuals involved in wetland and other resource inventories, wetland identification and delineation, and wetland research. Interim Regional Interagency Review Panel (Regional Panel) changes in indicator status as well as additions and deletions to the 1988 National List were documented in Regional supplements. The National List was originally developed as an appendix to the Classification of Wetlands and Deepwater Habitats of the United States (Cowardin et al.1979) to aid in the consistent application of this classification system for wetlands in the field.. The 1996 National List also was developed to aid in determining the presence of hydrophytic vegetation in the Clean Water Act Section 404 wetland regulatory program and in the implementation of the swampbuster provisions of the Food Security Act. While not required by law or regulation, the Fish and Wildlife Service is making the 1996 National List available for review and comment. -

DISSERTAÇÃO Caroline Leal Rodrigues Soares.Pdf
UNIVERSIDADE FEDERAL DE PERNAMBUCO CENTRO DE BIOCIÊNCIAS PROGRAMA DE PÓS GRADUAÇÃO EM CIÊNCIAS BIOLÓGICAS CAROLINE LEAL RODRIGUES SOARES AVALIAÇÃO DA ATIVIDADE CICATRIZANTE IN VITRO E IN VIVO DO EXTRATO HIDROALCOÓLICO DE LAGUNCULARIA RACEMOSA (L) C.F. GAERTN Recife 2018 CAROLINE LEAL RODRIGUES SOARES AVALIAÇÃO DA ATIVIDADE CICATRIZANTE IN VITRO E IN VIVO DO EXTRATO HIDROALCOÓLICO DE LAGUNCULARIA RACEMOSA (L) C.F. GAERTN Dissertação apresentada ao Programa de Pós- Graduação em Ciências Biológicas da Universidade Federal de Pernambuco, como parte dos requisitos parciais para obtenção do título de mestre em Ciências Biológicas. Área de concentração: Biologia Química para a Saúde Orientadora: Profª. Drª. Teresinha Gonçalves da Silva Recife 2018 Dados Internacionais de Catalogação na Publicação (CIP) de acordo com ISBD Soares, Caroline Leal Rodrigues Avaliação da atividade cicatrizante in vitro e in vivo do extrato hidroalcoólico de Laguncularia racemosa (L) C.F. Gaertn / Caroline Leal Rodrigues Soares - 2018. 85 folhas: il., fig., tab. Orientadora: Teresinha Gonçalves da Silva Dissertação (Mestrado) – Universidade Federal de Pernambuco. Centro de Biociências. Programa de Pós-Graduação em Ciências Biológicas. Recife, 2018. Inclui referências e anexo 1. Plantas medicinais 2. Laguncularia racemosa 3. Cicatrização I. Silva, Teresinha Gonçalves da (orient.) II. Título 615.321 CDD (22.ed.) UFPE/CB-2018-452 Elaborado por Claudina Karla Queiroz Ribeiro CRB4/1752 CAROLINE LEAL RODRIGUES SOARES AVALIAÇÃO DA ATIVIDADE CICATRIZANTE IN VITRO E IN VIVO DO EXTRATO HIDROALCOÓLICO DE LAGUNCULARIA RACEMOSA (L) C.F. GAERTN Dissertação apresentada ao Programa de Pós- Graduação em Ciências Biológicas da Universidade Federal de Pernambuco, como parte dos requisitos parciais para obtenção do título de mestre em Ciências Biológicas. -

Leaf Morphological Strategies of Seedlings and Saplings Of
Leaf morphological strategies of seedlings and saplings of Rhizophora mangle (Rhizophoraceae), Laguncularia racemosa (Combretaceae) and Avicennia schaueriana (Acanthaceae) from Southern Brazil Andressa Pelozo1, Maria Regina T. Boeger2, Carolina Sereneski-de-Lima3 & Patricia Soffiatti4* 1. Universidade Federal do Paraná, Programa de Pós-Graduação em Botânica, Departamento de Botânica, SCB, Centro Politécnico, Caixa Postal 19031, Curitiba, PR. Brazil, CEP 81.531-880; [email protected] 2. Programa de Pós-Graduação em Ecologia, SCB, Centro Politécnico, Caixa Postal 19031, Curitiba, PR, Brazil, CEP 81.531-880; [email protected] 3. Programa de Pós-Graduação em Ecologia, SCB, Centro Politécnico, Caixa Postal 19031, Curitiba, PR, Brazil, CEP 81.531-880; [email protected] 4. Universidade Federal do Paraná, Departamento de Botânica, SCB, Centro Politécnico, Caixa Postal 19031, Curitiba, PR. Brazil, CEP 81.531-880; [email protected] * Correspondence Received 26-I-2015. Corrected 27-VII-2015. Accepted 25-VIII-2015. Abstract: The initial phase of a plant life cycle is a short and critical period, when individuals are more vul- nerable to environmental factors. The morphological and anatomical study of seedlings and saplings leaf type enables the understanding of species strategies of fundamental importance in their establishment and survival. The objective of this study was to analyze the structure of seedlings and saplings leaf types of three mangrove species, Avicennia schaueriana, Laguncularia racemosa, Rhizophora mangle, to understand their early life adap- tive strategies to the environment. A total of 30 fully expanded cotyledons (A. schaueriana and L. racemosa), 30 leaves of seedlings, and 30 leaves of saplings of each species were collected from a mangrove area in Guaratuba Bay, Paraná State, Brazil. -

Fl. China 19: 218–220. 2011. 53. MITRAGYNA Korthals, Observ. Naucl. Indic. 19. 1839, Nom. Cons., Not Mitragyne R. Brown (1810
Fl. China 19: 218–220. 2011. 53. MITRAGYNA Korthals, Observ. Naucl. Indic. 19. 1839, nom. cons., not Mitragyne R. Brown (1810). 帽蕊木属 mao rui mu shu Chen Tao (陈涛); Charlotte M. Taylor Paradina Pierre ex Pitard; Stephegyne Korthals. Trees, unarmed; buds flattened, with stipules erect and pressed together. Raphides absent. Leaves opposite, sometimes with domatia; stipules caducous, interpetiolar, generally ovate to obovate, sometimes keeled, entire, often well developed. Inflorescences terminal on main stems and axillary branches and often accompanied by reduced, petaloid, and/or bracteate leaves, capitate with globose heads in fascicles, cymes, umbels, or thyrses, sessile to shortly pedunculate, bracteate; bracteoles spatulate to obpyramidal. Flowers sessile, bisexual, monomorphic. Calyx limb truncate to 5-lobed. Corolla cream to yellow-green, funnelform or narrowly salverform, inside glabrous to variously pubescent; lobes 5, valvate in bud. Stamens 5, inserted near corolla throat, exserted or included; filaments short; anthers basifixed, partially to fully exserted. Ovary 2-celled, ovules numerous in each cell on fleshy, pendulous, axile placentas attached in upper third of septum; stigma clavate to mitriform (i.e., upside-down cupular), exserted. Fruit capsular, obovoid to ellipsoid, septicidally then loculicidally dehiscent, cartilaginous to woody, with calyx limb persistent or decidu- ous; seeds numerous, small, somewhat flattened, fusiform to lanceolate, shortly winged at both ends with basal wing sometimes bifid or notched. About seven species: one species in Africa, six species in Asia and Malesia; three species in China. Ridsdale reviewed this genus in detail (Blumea 24: 46–68. 1978) and excluded the African species. H. H. Hsue and H. Wu (in FRPS 71(1): 245. -

National List of Vascular Plant Species That Occur in Wetlands 1996
National List of Vascular Plant Species that Occur in Wetlands: 1996 National Summary Indicator by Region and Subregion Scientific Name/ North North Central South Inter- National Subregion Northeast Southeast Central Plains Plains Plains Southwest mountain Northwest California Alaska Caribbean Hawaii Indicator Range Abies amabilis (Dougl. ex Loud.) Dougl. ex Forbes FACU FACU UPL UPL,FACU Abies balsamea (L.) P. Mill. FAC FACW FAC,FACW Abies concolor (Gord. & Glend.) Lindl. ex Hildebr. NI NI NI NI NI UPL UPL Abies fraseri (Pursh) Poir. FACU FACU FACU Abies grandis (Dougl. ex D. Don) Lindl. FACU-* NI FACU-* Abies lasiocarpa (Hook.) Nutt. NI NI FACU+ FACU- FACU FAC UPL UPL,FAC Abies magnifica A. Murr. NI UPL NI FACU UPL,FACU Abildgaardia ovata (Burm. f.) Kral FACW+ FAC+ FAC+,FACW+ Abutilon theophrasti Medik. UPL FACU- FACU- UPL UPL UPL UPL UPL NI NI UPL,FACU- Acacia choriophylla Benth. FAC* FAC* Acacia farnesiana (L.) Willd. FACU NI NI* NI NI FACU Acacia greggii Gray UPL UPL FACU FACU UPL,FACU Acacia macracantha Humb. & Bonpl. ex Willd. NI FAC FAC Acacia minuta ssp. minuta (M.E. Jones) Beauchamp FACU FACU Acaena exigua Gray OBL OBL Acalypha bisetosa Bertol. ex Spreng. FACW FACW Acalypha virginica L. FACU- FACU- FAC- FACU- FACU- FACU* FACU-,FAC- Acalypha virginica var. rhomboidea (Raf.) Cooperrider FACU- FAC- FACU FACU- FACU- FACU* FACU-,FAC- Acanthocereus tetragonus (L.) Humm. FAC* NI NI FAC* Acanthomintha ilicifolia (Gray) Gray FAC* FAC* Acanthus ebracteatus Vahl OBL OBL Acer circinatum Pursh FAC- FAC NI FAC-,FAC Acer glabrum Torr. FAC FAC FAC FACU FACU* FAC FACU FACU*,FAC Acer grandidentatum Nutt. -

MANILA BAY AREA SITUATION ATLAS December 2018
Republic of the Philippines National Economic and Development Authority Manila Bay Sustainable Development Master Plan MANILA BAY AREA SITUATION ATLAS December 2018 MANILA BAY AREA SITUATION ATLAS December 2018 i Table of Contents Preface, v Administrative and Institutional Systems, 78 Introduction, 1 Administrative Boundaries, 79 Natural Resources Systems, 6 Stakeholders Profile, 85 Climate, 7 Institutional Setup, 87 Topography, 11 Public-Private Partnership, 89 Geology, 13 Budget and Financing, 91 Pedology, 15 Policy and Legal Frameworks, 94 Hydrology, 17 National Legal Framework, 95 Oceanography, 19 Mandamus Agencies, 105 Land Cover, 21 Infrastructure, 110 Hazard Prone Areas, 23 Transport, 111 Ecosystems, 29 Energy, 115 Socio-Economic Systems, 36 Water Supply, 119 Population and Demography, 37 Sanitation and Sewerage, 121 Settlements, 45 Land Reclamation, 123 Waste, 47 Shoreline Protection, 125 Economics, 51 State of Manila Bay, 128 Livelihood and Income, 55 Water Quality Degradation, 129 Education and Health, 57 Air Quality, 133 Culture and Heritage, 61 Habitat Degradation, 135 Resource Use and Conservation, 64 Biodiversity Loss, 137 Agriculture and Livestock, 65 Vulnerability and Risk, 139 Aquaculture and Fisheries, 67 References, 146 Tourism, 73 Ports and Shipping, 75 ii Acronyms ADB Asian Development Bank ISF Informal Settlers NSSMP National Sewerage and Septage Management Program AHLP Affordable Housing Loan Program IUCN International Union for Conservation of Nature NSWMC National Solid Waste Management Commission AQI Air Quality Index JICA Japan International Cooperation Agency OCL Omnibus Commitment Line ASEAN Association of Southeast Nations KWFR Kaliwa Watershed Forest Reserve OECD Organization for Economic Cooperation and Development BSWM Bureau of Soils and Water Management LGU Local Government Unit OIDCI Orient Integrated Development Consultants, Inc. -
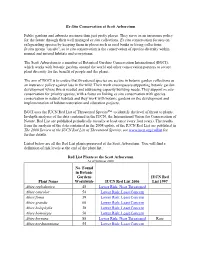
IUCN Red List of Threatened Species™ to Identify the Level of Threat to Plants
Ex-Situ Conservation at Scott Arboretum Public gardens and arboreta are more than just pretty places. They serve as an insurance policy for the future through their well managed ex situ collections. Ex situ conservation focuses on safeguarding species by keeping them in places such as seed banks or living collections. In situ means "on site", so in situ conservation is the conservation of species diversity within normal and natural habitats and ecosystems. The Scott Arboretum is a member of Botanical Gardens Conservation International (BGCI), which works with botanic gardens around the world and other conservation partners to secure plant diversity for the benefit of people and the planet. The aim of BGCI is to ensure that threatened species are secure in botanic garden collections as an insurance policy against loss in the wild. Their work encompasses supporting botanic garden development where this is needed and addressing capacity building needs. They support ex situ conservation for priority species, with a focus on linking ex situ conservation with species conservation in natural habitats and they work with botanic gardens on the development and implementation of habitat restoration and education projects. BGCI uses the IUCN Red List of Threatened Species™ to identify the level of threat to plants. In-depth analyses of the data contained in the IUCN, the International Union for Conservation of Nature, Red List are published periodically (usually at least once every four years). The results from the analysis of the data contained in the 2008 update of the IUCN Red List are published in The 2008 Review of the IUCN Red List of Threatened Species; see www.iucn.org/redlist for further details. -

Differentiation of Mitragyna Speciosa, a Narcotic Plant, from Allied Mitragyna Species Using DNA Barcoding-High-Resolution Melting (Bar-HRM) Analysis
Differentiation of Mitragyna Speciosa, a Narcotic Plant, From Allied Mitragyna Species Using DNA Barcoding-high-resolution Melting (Bar-HRM) Analysis Chayapol Tungphatthong Chulalongkorn University Santhosh Kumar J Urumarudappa Chulalongkorn University Supita Awachai Chulalongkorn University Thongchai Sooksawate Chulalongkorn University Suchada Sukrong ( [email protected] ) Chulalongkorn University Research Article Keywords: Mitragyna speciosa, Kratom, Rubiaceae, Bar-HRM, Narcotic plant Posted Date: November 25th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-109985/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published at Scientic Reports on March 24th, 2021. See the published version at https://doi.org/10.1038/s41598-021-86228-9. Page 1/16 Abstract Mitragyna speciosa (Korth.) Havil. [MS], or “kratom” in Thai, is the only narcotic species among the four species of Mitragyna in Thailand, which also include Mitragyna diversifolia (Wall. ex G. Don) Havil. [MD], Mitragyna hirsuta Havil. [MH], and Mitragyna rotundifolia (Roxb.) O. Kuntze [MR]. M. speciosa is a tropical tree belonging to the Rubiaceae family and has been prohibited by law in Thailand. However, it has been extensively covered in national and international news, as its abuse has become more popular. M. speciosa is a narcotic plant and has been used as an opium substitute and traditionally used for the treatment of chronic pain and various illnesses. Due to morphological disparities in the genus, the identication of plants in various forms, including fresh leaves, dried leaf powder, and nished products, is dicult. In this study, DNA barcoding combined with high-resolution melting (Bar-HRM) analysis was performed to differentiate M. -

John Day Fossil Beds NM: Geology and Paleoenvironments of the Clarno Unit
John Day Fossil Beds NM: Geology and Paleoenvironments of the Clarno Unit JOHN DAY FOSSIL BEDS Geology and Paleoenvironments of the Clarno Unit John Day Fossil Beds National Monument, Oregon GEOLOGY AND PALEOENVIRONMENTS OF THE CLARNO UNIT John Day Fossil Beds National Monument, Oregon By Erick A. Bestland, PhD Erick Bestland and Associates, 1010 Monroe St., Eugene, OR 97402 Gregory J. Retallack, PhD Department of Geological Sciences University of Oregon Eugene, OR 7403-1272 June 28, 1994 Final Report NPS Contract CX-9000-1-10009 TABLE OF CONTENTS joda/bestland-retallack1/index.htm Last Updated: 21-Aug-2007 http://www.nps.gov/history/history/online_books/joda/bestland-retallack1/index.htm[4/18/2014 12:20:25 PM] John Day Fossil Beds NM: Geology and Paleoenvironments of the Clarno Unit (Table of Contents) JOHN DAY FOSSIL BEDS Geology and Paleoenvironments of the Clarno Unit John Day Fossil Beds National Monument, Oregon TABLE OF CONTENTS COVER ABSTRACT ACKNOWLEDGEMENTS CHAPTER I: INTRODUCTION AND REGIONAL GEOLOGY INTRODUCTION PREVIOUS WORK AND REGIONAL GEOLOGY Basement rocks Clarno Formation John Day Formation CHAPTER II: GEOLOGIC FRAMEWORK INTRODUCTION Stratigraphic nomenclature Radiometric age determinations CLARNO FORMATION LITHOSTRATIGRAPHIC UNITS Lower Clarno Formation units Main section JOHN DAY FORMATION LITHOSTRATIGRAPHIC UNITS Lower Big Basin Member Middle and upper Big Basin Member Turtle Cove Member GEOCHEMISTRY OF LAVA FLOW AND TUFF UNITS Basaltic lava flows Geochemistry of andesitic units Geochemistry of tuffs STRUCTURE OF CLARNO -

Chloroplast Genome Analysis of Angiosperms and Phylogenetic Relationships Among
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.05.078212; this version posted May 5, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Chloroplast genome analysis of Angiosperms and phylogenetic relationships among Lamiaceae members with particular reference to teak (Tectona grandis L.f) P. MAHESWARI, C. KUNHIKANNAN AND R. YASODHA* Institute of Forest Genetics and Tree Breeding, Coimbatore 641 002 INDIA *Author for correspondence R. YASODHA, Institute of Forest Genetics and Tree Breeding, Coimbatore, India Telephone: +91 422 2484114; Fax number : +91 422 248549; e.mail: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.05.05.078212; this version posted May 5, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Availability of comprehensive phylogenetic tree for flowering plants which includes many of the economically important crops and trees is one of the essential requirements of plant biologists for diverse applications. It is the first study on the use of chloroplast genome of 3265 Angiosperm taxa to identify evolutionary relationships among the plant species. Sixty genes from chloroplast genome was concatenated and utilized to generate the phylogenetic tree. Overall the phylogeny was in correspondence with Angiosperm Phylogeny Group (APG) IV classification with very few taxa occupying incongruous position either due to ambiguous taxonomy or incorrect identification. Simple sequence repeats (SSRs) were identified from almost all the taxa indicating the possibility of their use in various genetic analyses. -

Fragrant Annuals Fragrant Annuals
TheThe AmericanAmerican GARDENERGARDENER® TheThe MagazineMagazine ofof thethe AAmericanmerican HorticulturalHorticultural SocietySociety JanuaryJanuary // FebruaryFebruary 20112011 New Plants for 2011 Unusual Trees with Garden Potential The AHS’s River Farm: A Center of Horticulture Fragrant Annuals Legacies assume many forms hether making estate plans, considering W year-end giving, honoring a loved one or planting a tree, the legacies of tomorrow are created today. Please remember the American Horticultural Society when making your estate and charitable giving plans. Together we can leave a legacy of a greener, healthier, more beautiful America. For more information on including the AHS in your estate planning and charitable giving, or to make a gift to honor or remember a loved one, please contact Courtney Capstack at (703) 768-5700 ext. 127. Making America a Nation of Gardeners, a Land of Gardens contents Volume 90, Number 1 . January / February 2011 FEATURES DEPARTMENTS 5 NOTES FROM RIVER FARM 6 MEMBERS’ FORUM 8 NEWS FROM THE AHS 2011 Seed Exchange catalog online for AHS members, new AHS Travel Study Program destinations, AHS forms partnership with Northeast garden symposium, registration open for 10th annual America in Bloom Contest, 2011 EPCOT International Flower & Garden Festival, Colonial Williamsburg Garden Symposium, TGOA-MGCA garden photography competition opens. 40 GARDEN SOLUTIONS Plant expert Scott Aker offers a holistic approach to solving common problems. 42 HOMEGROWN HARVEST page 28 Easy-to-grow parsley. 44 GARDENER’S NOTEBOOK Enlightened ways to NEW PLANTS FOR 2011 BY JANE BERGER 12 control powdery mildew, Edible, compact, upright, and colorful are the themes of this beating bugs with plant year’s new plant introductions. -

The Chinaberry Path with Melia Azedarach Flowering Profusely At
photograph © Jim Ethridge The Chinaberry path with Melia azedarach flowering profusely at Quarryhill Botanical Garden in California, which celebrated its twenty-fifth anniversary in 2012. photograph © Tony Kirkham The view across the valley from the prayer flags at Quarryhill. 138 Quarryhill Botanical Garden, USA In 1987 Jane Davenport Hansen started creating the botanical garden that is Quarryhill in Sonoma Valley, California. TONY KIRKHAM writes about the exceptional collection of wild collected Chinese plants that thrive there 25 years on. 1987 can be remembered in the Dendrological calendar for a number of events in the tree world, particularly the Great Storm in the South-east of England that destroyed over 15 million trees in one night. This storm was a real wakeup call to anyone with even a slight interest in trees and it started a new beginning in the tree world, raising public awareness and generating new planting schemes across the UK. In the same year, in Sonoma Valley, California, Jane Davenport Jansen had a vision that would make significant additions to arboreta across the botanic garden world, and leave a living legacy that is now one of the largest living collections of wild collected temperate Asian plants in North America; it was the creation of Quarryhill Botanical Garden. In 1968, Jane purchased 61 acres in the foothills of the Mayacama Mountains off the Sonoma Highway in Glen Ellen, Sonoma County to be planted with vineyards, but 20 acres of the hillside contained several abandoned stone quarries and in 1987 she decided to create a garden. Unlike many gardens INTERNATIONAL DENDROLOGY SOCIETY GARDENS & ARBORETA that would be planted purely for landscape, aesthetic and floral qualities, Jane had set her sights on something unique to the area that would become her personal drive, a landscape planted with natural source plants derived from her own funded expeditions to targeted countries around the world with an emphasis on the Far East.