The Role of Pharmacogenomics in Improving the Management of Asthma
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Drug Information Sheet("Kusuri-No-Shiori")
Drug Information Sheet("Kusuri-no-Shiori") Internal Published: 05/2017 The information on this sheet is based on approvals granted by the Japanese regulatory authority. Approval details may vary by country. Medicines have adverse reactions (risks) as well as efficacies (benefits). It is important to minimize adverse reactions and maximize efficacy. To obtain a better therapeutic response, patients should understand their medication and cooperate with the treatment. Brand name:PRANLUKAST TABLETS 112.5mg "CEO" Active ingredient:Pranlukast hydrate Dosage form:white to pale yellow tablet, diameter: 7.5 mm, thickness: 2.7 mm Print on wrapping:(face) プランルカスト 112.5mg「CEO」, CEO 131, プランルカスト 112.5mg (back) PRANLUKAST 112.5mg「CEO」, CEO 131, プランルカスト 112.5mg Effects of this medicine This medicine selectively binds to leukotriene receptor and inhibits its action. It consequently suppresses increase in airway contraction, vascular permeability, mucosal edema and hypersensitivity. It is usually used to treat bronchial asthma and allergic rhinitis. However, it cannot stop the attack of bronchial asthma already in progress but prevents the asthma attack. Before using this medicine, be sure to tell your doctor and pharmacist ・If you have previously experienced any allergic reactions (itch, rash, etc.) to any medicines. ・If you are pregnant or breastfeeding. ・If you are taking any other medicinal products. (Some medicines may interact to enhance or diminish medicinal effects. Beware of over-the-counter medicines and dietary supplements as well as other prescription medicines.) Dosing schedule (How to take this medicine) ・Your dosing schedule prescribed by your doctor is(( to be written by a healthcare professional)) ・In general, for adults, take 2 tablets (225 mg of the active ingredient) at a time, twice a day after breakfast and dinner. -

Inhibitory Activity of Pranlukast and Montelukas Against Histamine
Showa Univ J Med Sci 21(2), 77~84, June 2009 Original Inhibitory Activity of Pranlukast and Montelukas Against Histamine Release and LTC4 Production from Human Basophils 1, 2 1 1 Satoshi HIBINO ), Ryoko ITO ), Taeru KITABAYASHI ), 1 2 Kazuo ITAHASHI ) and Toshio NAKADATE ) Abstract : Leukotriene receptor antagonists(LTRAs)are routinely used to treat bronchial asthma and are thought to act mostly by inhibiting leukotriene receptors. However, there is no preclinical or clinical evidence of the direct effect of LTRAs on histamine release from and leukotriene(LT)C4 produc- tion by basophils. We used anti-IgE antibody(Ab), FMLP, and C5a to induce histamine release, and anti-IgE Ab and FMLP to stimulate LTC 4 production. Basophils were exposed to different concentrations of pranlukast and montelu- kast, and then to anti-IgE Ab, FMLP, and C5a. Culture supernatant histamine and LTC 4 levels were measured by using a histamine ELISA kit and a LTC 4 EIA kit, respectively. Histamine release was expressed as a percentage of the total histamine content(%HR)induced by anti-IgE Ab, FMLP, or C5a. To evaluate the effects of pranlukast and montelukast on histamine release and LTC 4 production, we calculated the percent inhibition of histamine release and LTC 4 production, expressed as percent inhibition, at different concentrations of pranlukast and montelukast. Pranlukast significantly inhibited histamine release stimulated by FMLP and C5a, but had no effect on histamine release stimulated by anti-IgE Ab. By comparison, montelukast signicantly inhibited histamine release stimulated by FMLP, C5a, and anti-IgE Ab, in a concentration-dependent manner. Both pranlukast and montelukast signicantly inhibited LTC 4 production stimulated by anti-IgE Ab and FMLP. -

Drug Class Review Controller Medications for Asthma
Drug Class Review Controller Medications for Asthma Final Update 1 Report April 2011 The Agency for Healthcare Research and Quality has not yet seen or approved this report. The purpose of this report is to make available information regarding the comparative effectiveness and safety profiles of different drugs within pharmaceutical classes. Reports are not usage guidelines, nor should they be read as an endorsement of, or recommendation for, any particular drug, use or approach. Oregon Health & Science University does not recommend or endorse any guideline or recommendation developed by users of these reports. Original Report: November 2008 Daniel E. Jonas, MD, MPH Roberta C. M. Wines, MPH Marcy DelMonte, PharmD, BCPS Halle R. Amick, MSPH Tania M. Wilkins, MS Brett D. Einerson, MPH Christine L. Schuler, MD Blake A. Wynia, MPH Betsy Bryant Shilliday, Pharm.D., CDE, CPP Produced by RTI-UNC Evidence-based Practice Center Cecil G. Sheps Center for Health Services Research University of North Carolina at Chapel Hill 725 Martin Luther King Jr. Blvd, CB# 7590 Chapel Hill, NC 27599-7590 Tim Carey, M.D., M.P.H., Director Oregon Evidence-based Practice Center Oregon Health & Science University Mark Helfand, MD, MPH, Director Copyright © 2011 by Oregon Health & Science University Portland, Oregon 97239. All rights reserved. Final Update 1 Report Drug Effectiveness Review Project The medical literature relating to this topic is scanned periodically. (See http://www.ohsu.edu/xd/research/centers-institutes/evidence-based-policy- center/derp/documents/methods.cfm for description of scanning process). Prior versions of this report can be accessed at the DERP website. -

Understanding Interparticle Interactions in Dry Powder Inhalation: Glass Beads As an Innovative Model Carrier System
UNDERSTANDING INTERPARTICLE INTERACTIONS IN DRY POWDER INHALATION: GLASS BEADS AS AN INNOVATIVE MODEL CARRIER SYSTEM DOCTORAL THESIS SUMBITTED IN FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR IN NATURAL SCIENCES AT KIEL UNIVERSITY, GERMANY by Niklas Ludwig Renner Kiel 2017 Referee: Prof. Dr. Regina Scherließ Co-Referee: Prof. Dr. Thomas Kunze Date of Exam: 13.10.2017 Accepted for publication: 13.10.2017 Prof. Dr. Natascha Oppelt (Dean) Published research articles contributing to the present thesis: Renner, N.; Steckel, H.; Urbanetz, N.A.; Scherließ, R. Tailoring the surface topography of a model carrier to alter dry powder inhaler performance Dalby, R.N. (Ed.), RDD Europe 2017 2 (2017), 195-200 Renner, N; Steckel, H.; Urbanetz, N.A.; Scherließ, R. Nano- and microstructured model carrier surfaces to alter dry powder inhaler performance International Journal of Pharmaceutics 518 (2017), 20-28 Conference contributions: Renner, N.; Steckel, H.; Urbanetz, N.A.; Scherließ, R. Tailoring the surface topography of a model carrier to alter dry powder inhaler per- formance Respiratory Drug Delivery, Nice, France (2017) Renner, N.; Steckel, H.; Urbanetz, N.A.; Scherließ, R. A deeper insight into the impact of chemical surface properties on inhalation perfor- mance Drug Delivery to the Lungs 27, Edinburgh, Scotland (2016) Renner, N.; Scherließ, R.; Steckel, H. Glass beads as model carriers in dry powder inhalers: the influence of chemical sur- face properties on inhalation performance International Congress on Particle Technology, Nurnberg, Germany (2016) Renner, N.; Scherließ, R.; Steckel, H. Investigating the influence of carrier surface roughness on drug delivery in DPIs 10th World Meeting on Pharmaceutics, Biopharmaceutics and Pharmaceutical Tech- nology, Glasgow, Scotland (2016) Renner, N.; Kutelova, Z.; Scherließ, R.; Steckel, H. -

The Role of 5-Lipoxygenase in Pathophysiology and Management of Neuropathic Pain
REVIEW ARTICLE THE ROLE OF 5-LIPOXYGENASE IN PATHOPHYSIOLOGY AND MANAGEMENT OF NEUROPATHIC PAIN Pascanus Lamsihar PT∗, Faldi Yaputra∗∗, Jimmy FA Barus4 and I Putu Eka Widyadharma∗∗,1 ∗Department of Neurology, Provincial Mental Hospital, West Borneo, Indonesia., ∗∗Department of Neurology, Faculty of Medicine, Udayana University-Sanglah General Hospital, Bali, Indonesia., 4Department of Neurology, Faculty of Medicine, Atma Jaya Catholic University, Jakarta-Indonesia. ABSTRACT Neuropathic pain (NP) is a pain caused by lesions in the nervous system. Several causes of NP are traumatic, metabolic disorders, ischemia, toxins, infections, immune-related, and hereditary. The pathophysiology of NP is very complicated and unknown entirely. Therefore the treatment of NP is still unsatisfactory. Recent studies believed the critical role of primary inflammatory mediators in the pathophysiology of NP especially leukotrienes (LTs). The 5- lipoxygenase enzyme (5-LOX) is an enzyme that plays a role in the metabolism of arachidonic acid into LTs. Leukotrienes (LTs) are the essential inflammatory mediators in the pathophysiology of NP. Leukotriene B4 (LTB4) can cause chemotaxis on neutrophils, lowering nociceptors threshold and may contribute to NP. Several studies believed the administration of 5-LOX inhibitors or LTs receptor antagonists could be useful in the management of NP. The purpose of this review is to summarize the involvement of 5-LOX enzyme as an essential role in the pathophysiology and management of NP. KEYWORDS 5-lipoxygenase, leukotrienes, -
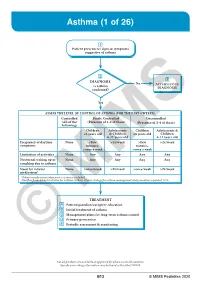
Asthma (1 of 26)
Asthma (1 of 26) 1 Patient presents w/ signs & symptoms suggestive of asthma 2 3 DIAGNOSIS No ALTERNATIVE Is asthma DIAGNOSIS confi rmed? Yes ASSESS THE LEVEL OF CONTROL OF ASTHMA FOR THE PAST 4 WEEKS Controlled Partly Controlled Uncontrolled (All of the (Presence of 1-2 of these) (Presence of 3-4 of these) following) Children Adolescents Children Adolescents & ≤5 years old & Children ≤5 years old Children 6-11 years old 6-11 years old Frequency of daytime None >Few >2x/week >Few >2x/week symptoms minutes, minutes, >once a week >once a week Limitation of activities None Any Any Any Any Nocturnal waking up or None Any Any Any Any coughing due to asthma Need for reliever None >once/week >2x/week >once/week >2x/week medication* *Reliever medications taken prior to exercise excluded. Modified from: Global Initiative for Asthma (GINA). Global strategy for asthma management and prevention: Updated 2020. TREATMENT A Patient/guardian/caregiver education B Initial treatment of asthma C Management plans for long-term asthma control D Primary prevention E © Periodic assessmentMIMS & monitoring Not all products are available or approved for above use in all countries. Specifi c prescribing information may be found in the latest MIMS. B13 © MIMS Pediatrics 2020 Asthma (2 of 26) 1 ASTHMA • A heterogeneous disease w/ chronic infl ammatory disorder of the airways • e most common chronic disease in pediatric age groups that causes signifi cant morbidity • Characterized by history of respiratory symptoms eg wheeze, shortness of breath, chest tightness & cough -

2020 GINA Pocket Guide for Asthma Management and Prevention
POCKET GUIDE FOR ASTHMA MANAGEMENT AND PREVENTION (for Adults and Children Older than 5 Years) DISTRIBUTE OR COPY NOT DO MATERIAL- COPYRIGHTED A Pocket Guide for Health Professionals Updated 2020 BASED ON THE GLOBAL STRATEGY FOR ASTHMA MANAGEMENT AND PREVENTION © 2020 Global Initiative for Asthma GLOBAL INITIATIVE FOR ASTHMA ASTHMA MANAGEMENT AND PREVENTION DISTRIBUTE for adults and children older ORthan 5 years COPY NOT A POCKET GUIDE FOR HEALTHDO PROFESSIONALS MATERIAL-Updated 2020 GINA Science Committee Chair: Helen Reddel, MBBS PhD COPYRIGHTED GINA Board of Directors Chair: Louis-Philippe Boulet, MD GINA Dissemination and Implementation Committee Chair: Mark Levy, MD (to Sept 2019); Alvaro Cruz, MD (from Sept 2019) GINA Assembly The GINA Assembly includes members from many countries, listed on the GINA website www.ginasthma.org. GINA Executive Director Rebecca Decker, BS, MSJ Names of members of the GINA Committees are listed on page 48. 1 LIST OF ABBREVIATIONS BDP Beclometasone dipropionate COPD Chronic obstructive pulmonary disease CXR Chest X-ray DPI Dry powder inhaler FeNO Fraction of exhaled nitric oxide FEV1 Forced expiratory volume in 1 second FVC Forced vital capacity GERD Gastroesophageal reflux disease HDM House dust mite ICS Inhaled corticosteroids ICS-LABA Combination ICS and LABA Ig Immunoglobulin IL Interleukin DISTRIBUTE IV Intravenous OR LABA Long-acting beta2-agonist COPY LAMA Long-acting muscarinic antagonist NOT LTRA Leukotriene receptor antagonistDO n.a. Not applicable NSAID Nonsteroidal anti-inflammatory drug MATERIAL- O2 Oxygen OCS Oral corticosteroids PEF Peak expiratory flow pMDI PressurizedCOPYRIGHTED metered dose inhaler SABA Short-acting beta2-agonist SC Subcutaneous SLIT Sublingual immunotherapy 2 TABLE OF CONTENTS List of abbreviations ........................................................................................ -

Inflammation, Cancer and Oxidative Lipoxygenase Activity Are Intimately Linked
Cancers 2014, 6, 1500-1521; doi:10.3390/cancers6031500 OPEN ACCESS cancers ISSN 2072-6694 www.mdpi.com/journal/cancers Review Inflammation, Cancer and Oxidative Lipoxygenase Activity are Intimately Linked Rosalina Wisastra and Frank J. Dekker * Pharmaceutical Gene Modulation, Groningen Research Institute of Pharmacy, University of Groningen, Antonius Deusinglaan 1, 9713 AV Groningen, The Netherlands; E-Mail: [email protected] * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +31-5-3638030; Fax: +31-5-3637953. Received: 16 April 2014; in revised form: 27 June 2014 / Accepted: 2 July 2014 / Published: 17 July 2014 Abstract: Cancer and inflammation are intimately linked due to specific oxidative processes in the tumor microenvironment. Lipoxygenases are a versatile class of oxidative enzymes involved in arachidonic acid metabolism. An increasing number of arachidonic acid metabolites is being discovered and apart from their classically recognized pro-inflammatory effects, anti-inflammatory effects are also being described in recent years. Interestingly, these lipid mediators are involved in activation of pro-inflammatory signal transduction pathways such as the nuclear factor κB (NF-κB) pathway, which illustrates the intimate link between lipid signaling and transcription factor activation. The identification of the role of arachidonic acid metabolites in several inflammatory diseases led to a significant drug discovery effort around arachidonic acid metabolizing enzymes. However, to date success in this area has been limited. This might be attributed to the lack of selectivity of the developed inhibitors and to a lack of detailed understanding of the functional roles of arachidonic acid metabolites in inflammatory responses and cancer. -

Antileukotriene Agents in Asthma: the Dart That Kills the Elephant?
Antileukotriene agents in asthma: The dart that kills the elephant? Education Paolo M. Renzi, MD Éducation Abstract From the Research Centre and the Pulmonary Unit, THE PERSISTENCE OF AIRWAY INFLAMMATION is believed to cause the mechanical Centre hospitalier de changes and symptoms of asthma. After decades of research, a new class of med- l’Université de Montréal ication has emerged that focuses on leukotrienes, mediators of inflammation. These Hospitals, Université de substances are potent inducers of bronchoconstriction, increased vascular perme- Montréal, and the Meakins ability and mucus production, and they potentiate the influx of inflammatory cells Christie Laboratories, McGill in the airways of patients with asthma. In this article the author reviews the devel- University, Montreal, Que. opment, mechanism of action, and clinical and toxic effects of the leukotriene syn- thesis inhibitors and receptor antagonists that are entering the North American market. These agents can decrease airway response to antigen, airway hyperre- This article has been peer reviewed. sponsiveness and exercise-induced asthma. They are also effective inhibitors of ASA-induced symptoms. Although few published studies are available, the an- CMAJ 1999;160:217-23 tileukotrienes seem almost as effective in the management of chronic asthma as ß See related article page 209 low-dose inhaled corticosteroids, and their use permits a decrease in the frequency of use or dose of corticosteroids. Further evaluation and clinical experience will determine the position of targeted inhibition of the leukotriene pathway in the treatment of asthma. Résumé ON CROIT QUE LA PERSISTANCE DE L’INFLAMMATION des voies respiratoires provoque les changements mécaniques et les symptômes de l’asthme. -

Concentration-Dependent Noncysteinyl Leukotriene Type 1 Receptor-Mediated Inhibitory Activity of Leukotriene Receptor Antagonist
Concentration-Dependent Noncysteinyl Leukotriene Type 1 Receptor-Mediated Inhibitory Activity of Leukotriene Receptor Antagonists This information is current as of September 27, 2021. Grzegorz Woszczek, Li-Yuan Chen, Sara Alsaaty, Sahrudaya Nagineni and James H. Shelhamer J Immunol 2010; 184:2219-2225; Prepublished online 18 January 2010; doi: 10.4049/jimmunol.0900071 Downloaded from http://www.jimmunol.org/content/184/4/2219 References This article cites 38 articles, 10 of which you can access for free at: http://www.jimmunol.org/ http://www.jimmunol.org/content/184/4/2219.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 27, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2010 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Concentration-Dependent Noncysteinyl Leukotriene Type 1 Receptor-Mediated Inhibitory Activity of Leukotriene Receptor Antagonists Grzegorz Woszczek,*,†,1 Li-Yuan Chen,*,1 Sara Alsaaty,* Sahrudaya Nagineni,* and James H. Shelhamer* The use of cysteinyl leukotriene receptor antagonists (LTRAs) for asthma therapy has been associated with a significant degree of interpatient variability in response to treatment. -

Pranlukast, a Novel Leukotriene Receptor Antagonist 525
Thorax 1997;52:523±527 523 Pranlukast, a novel leukotriene receptor Thorax: first published as 10.1136/thx.52.6.523 on 1 June 1997. Downloaded from antagonist: results of the ®rst European, placebo controlled, multicentre clinical study in asthma N C Barnes, J-C Pujet on behalf of an International Study Group Abstract asthma. It increased FEV1 within one hour Background ± Leukotriene receptor ant- of dosing, improved patient summary agonists have been shown to protect symptom and night-time asthma scores, against bronchoconstriction induced by and reduced the use of rescue broncho- antigens, exercise, and cold air. There are dilators, thus providing further evidence relatively few clinical studies reported in of a role for leukotrienes in the patho- patients with asthma. The present study genesis of asthma. is the ®rst clinical evaluation of pranlukast (Thorax 1997;52:523±527) (SB 205312, ONO-1078) outside Japan in patients with asthma. Keywords: leukotriene receptor antagonist, asthma, Methods ± A randomised, double blind, pranlukast, SB 205312, ONO-1078. placebo controlled, parallel group, multi- centre four week study of the safety and tolerability of oral pranlukast, 225 or Since the structure of leukotrienes was de- 337.5 mg twice daily, was performed in scribed in 1979,1 considerable eVort has been patients with mild to moderate asthma. made to develop drugs that block the leuko- Preliminary eYcacy data were obtained; triene receptor or inhibit synthesis of leuko- the main eYcacy variables evaluated were trienes. Challenge studies using a number forced expiratory volume in one second of leukotriene receptor antagonists or 5-lip- http://thorax.bmj.com/ (FEV1) and morning domiciliary (home) oxygenase inhibitors have shown that leuko- peak expiratory ¯ow rates (PEFR). -

Montelukast Versus Fluticasone: Effects on Lung Function, Airway
Copyright #ERS Journals Ltd 2002 Eur Respir J 2002; 20: 853–858 European Respiratory Journal DOI: 10.1183/09031936.02.00244602 ISSN 0903-1936 Printed in UK – all rights reserved Montelukast versus fluticasone: effects on lung function, airway responsiveness and inflammation in moderate asthma F. Kanniess, K. Richter, S. Bo¨hme, R.A. Jo¨rres, H. Magnussen Montelukast versus fluticasone: effects on lung function, airway responsiveness and Pulmonary Research Institute, Hospi- inflammation in moderate asthma. F. Kanniess, K. Richter, S. Bo¨hme, R.A. Jo¨rres, tal Grosshansdorf, Center for Pneu- H. Magnussen. #ERS Journals Ltd 2002. mology and Thoracic Surgery, D-22927 ABSTRACT: Whether leukotriene receptor antagonists exhibit adequate anti- Grosshansdorf, Germany. inflammatory effects in the treatment of asthma is still a controversial issue. The aim Correspondence: H. Magnussen, Pul- of the present study was to perform a direct comparison of the effects of a 4-week monary Research Institute, Hospital treatment with either montelukast (10 mg, once a day) or low-dose inhaled fluticasone Grosshansdorf, Center for Pneumology (100 mg b.i.d.) on functional and inflammatory parameters in steroid-naı¨ve patients with and Thoracic Surgery, Wo¨hrendamm moderate asthma. 80, D-22927 Grosshansdorf, Germany. Forty patients (forced expiratory volume in one second (FEV1), 60–80% predicted) Fax: 49 4102601379 were studied in a double-blind, randomised, crossover design. Treatment periods were E-mail: [email protected] separated by 3–8 weeks of washout. At the beginning and end of each period, FEV1, Keywords: Bronchial hyperresponsive- airway responsiveness to inhaled methacholine (provocative concentration causing a ness 20% fall in FEV1 (PC20)), the level of exhaled nitric oxide (NO) and sputum exhaled nitric oxide differential cell counts were determined.