Pymol Part 3
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Nano-Visualization Software for Education and Research
A Nano-Visualization Software for Education and Research Lillian C. Oetting Department of Computer Science, Stanford University, Stanford, CA 94305 USA West High School, Iowa City, IA 52246 USA Tehseen Z. Raza Department of Physics and Astronomy, University of Iowa, Iowa City, IA 52242 USA Hassan Raza Department of Electrical and Computer Engineering, University of Iowa, Iowa City, IA 52242 USA Centre for Fundamental Research, Islamabad, Pakistan Abstract: We report the development of a user-friendly nano-visualization software program which can acquaint high-school students with nanotechnology. The visual introduction to atoms and molecules, which are the building blocks of this technology, is an effective way to introduce the key concepts in this area. The software’s graphical user interface enables multidimensional atomic visualization by using ball and stick schematics. Additionally, the software provides the option of wavefunction visualization for arbitrary nanomaterials and nanostructures by using extended Hückel theory. The software is instructive, application oriented and may be useful not only in high school education but also for the undergraduate research and teaching. 1 I. Introduction: The ability to accurately depict atomic, molecular and electronic structures has been a key factor in the advancement of nanotechnology. In this context, it is imperative to provide teaching and research platforms to motivate students towards this novel technology [1-3], while keeping the societal implications in perspective [4]. Nano-visualization appeals well due to its simplistic, yet elegant approach towards the visual representation of detailed concepts about quantum mechanics, quantum chemistry and linear algebra. Additionally, the conflux of quantum mechanics, numerical computation, graphical design, and computer programming gives exposure to the multi-disciplinary aspect of this technology [5-8]. -

Python Tools in Computational Chemistry (And Biology)
Python Tools in Computational Chemistry (and Biology) Andrew Dalke Dalke Scientific, AB Göteborg, Sweden EuroSciPy, 26-27 July, 2008 “Why does ‘import numpy’ take 0.4 seconds? Does it need to import 228 libraries?” - My first Numpy-discussion post (paraphrased) Your use case isn't so typical and so suffers on the import time end of the balance. - Response from Robert Kern (Others did complain. Import time down to 0.28s.) 52,000 structures PDB doubles every 2½ years HEADER PHOTORECEPTOR 23-MAY-90 1BRD 1BRD 2 COMPND BACTERIORHODOPSIN 1BRD 3 SOURCE (HALOBACTERIUM $HALOBIUM) 1BRD 4 EXPDTA ELECTRON DIFFRACTION 1BRD 5 AUTHOR R.HENDERSON,J.M.BALDWIN,T.A.CESKA,F.ZEMLIN,E.BECKMANN, 1BRD 6 AUTHOR 2 K.H.DOWNING 1BRD 7 REVDAT 3 15-JAN-93 1BRDB 1 SEQRES 1BRDB 1 REVDAT 2 15-JUL-91 1BRDA 1 REMARK 1BRDA 1 .. ATOM 54 N PRO 8 20.397 -15.569 -13.739 1.00 20.00 1BRD 136 ATOM 55 CA PRO 8 21.592 -15.444 -12.900 1.00 20.00 1BRD 137 ATOM 56 C PRO 8 21.359 -15.206 -11.424 1.00 20.00 1BRD 138 ATOM 57 O PRO 8 21.904 -15.930 -10.563 1.00 20.00 1BRD 139 ATOM 58 CB PRO 8 22.367 -14.319 -13.591 1.00 20.00 1BRD 140 ATOM 59 CG PRO 8 22.089 -14.564 -15.053 1.00 20.00 1BRD 141 ATOM 60 CD PRO 8 20.647 -15.054 -15.103 1.00 20.00 1BRD 142 ATOM 61 N GLU 9 20.562 -14.211 -11.095 1.00 20.00 1BRD 143 ATOM 62 CA GLU 9 20.192 -13.808 -9.737 1.00 20.00 1BRD 144 ATOM 63 C GLU 9 19.567 -14.935 -8.932 1.00 20.00 1BRD 145 ATOM 64 O GLU 9 19.815 -15.104 -7.724 1.00 20.00 1BRD 146 ATOM 65 CB GLU 9 19.248 -12.591 -9.820 1.00 99.00 1 1BRD 147 ATOM 66 CG GLU 9 19.902 -11.351 -10.387 1.00 99.00 1 1BRD 148 ATOM 67 CD GLU 9 19.243 -10.169 -10.980 1.00 99.00 1 1BRD 149 ATOM 68 OE1 GLU 9 18.323 -10.191 -11.782 1.00 99.00 1 1BRD 150 ATOM 69 OE2 GLU 9 19.760 -9.089 -10.597 1.00 99.00 1 1BRD 151 ATOM 70 N TRP 10 18.764 -15.737 -9.597 1.00 20.00 1BRD 152 ATOM 71 CA TRP 10 18.034 -16.884 -9.090 1.00 20.00 1BRD 153 ATOM 72 C TRP 10 18.843 -17.908 -8.318 1.00 20.00 1BRD 154 ATOM 73 O TRP 10 18.376 -18.310 -7.230 1.00 20.00 1BRD 155 . -

Journal of Molecular Graphics and Modelling 73 (2017) 179–190
Journal of Molecular Graphics and Modelling 73 (2017) 179–190 Contents lists available at ScienceDirect Journal of Molecular Graphics and Modelling journa l homepage: www.elsevier.com/locate/JMGM Combined Monte Carlo/torsion-angle molecular dynamics for ensemble modeling of proteins, nucleic acids and carbohydrates a b c d,1 Weihong Zhang , Steven C. Howell , David W. Wright , Andrew Heindel , e a, b, Xiangyun Qiu , Jianhan Chen ∗, Joseph E. Curtis ∗∗ a Kansas State University, Manhattan, KS, USA b NIST Center for Neutron Research, 100 Bureau Drive, Gaithersburg, MD, USA c Centre for Computational Science, Department of Chemistry, University College London, 10 Gordon St., London, UK d James Madison University, Department of Chemistry & Biochemistry, Harrisonburg, VA, USA e Department of Physics, The George Washington University, Washington, DC, USA a r t i c l e i n f o a b s t r a c t Article history: We describe a general method to use Monte Carlo simulation followed by torsion-angle molecular dynam- Received 13 October 2016 ics simulations to create ensembles of structures to model a wide variety of soft-matter biological systems. Received in revised form 19 January 2017 Our particular emphasis is focused on modeling low-resolution small-angle scattering and reflectivity Accepted 17 February 2017 structural data. We provide examples of this method applied to HIV-1 Gag protein and derived fragment Available online 23 February 2017 proteins, TraI protein, linear B-DNA, a nucleosome core particle, and a glycosylated monoclonal antibody. This procedure will enable a large community of researchers to model low-resolution experimental data MSC: with greater accuracy by using robust physics based simulation and sampling methods which are a 00-01 99-00 significant improvement over traditional methods used to interpret such data. -

Biological Sciences 1
Biological Sciences 1 BIOL UN3208 Introduction to Evolutionary Biology or EEEB UN2001 BIOLOGICAL SCIENCES Environmental Biology I: Elements to Organisms. Departmental Office: 600 Fairchild, 212-854-4581; Interested students should consult listings in other departments for [email protected]; [email protected] courses related to biology. For courses in environmental studies, see listings for Earth and environmental sciences or for ecology, evolution, Director of Undergraduate Studies, Undergraduate Programs and and environmental biology. For courses in human evolution, see listings Laboratories: for anthropology or for ecology, evolution, and environmental biology. Prof. Deborah Mowshowitz, 744D Mudd; 212-854-4497; For courses in the history of evolution, see listings for history and for [email protected] philosophy of science. For a list of courses in computational biology and genomics, visit http://systemsbiology.columbia.edu/courses. Biology Major and Concentration Advisers: For a list of current biology, biochemistry, biophysics, and neuroscience and behavior advisers, please visit http://biology.columbia.edu/ Advanced Placement programs/advisors Transfer Credit A-F: Prof. Alice Heicklen, 744B Mudd; [email protected] Transfer credits granted toward the degree are not automatically counted G-O: Prof. Mary Ann Price, 744A Mudd; [email protected] toward the major. The department determines which transfer credits P-Z: Prof. Tulle Hazelrigg, 753A Mudd, [email protected] can be counted toward the major. For most majors, at least four biology Backup Advisor: Prof. Deborah Mowshowitz, 744D Mudd; 212-854-4497; or biochemistry courses and at least 18 credits of the total (biology, [email protected] biochemistry, math, physics, and chemistry) must be taken at Columbia. -

Chemdoodle Web Components: HTML5 Toolkit for Chemical Graphics, Interfaces, and Informatics Melanie C Burger1,2*
Burger. J Cheminform (2015) 7:35 DOI 10.1186/s13321-015-0085-3 REVIEW Open Access ChemDoodle Web Components: HTML5 toolkit for chemical graphics, interfaces, and informatics Melanie C Burger1,2* Abstract ChemDoodle Web Components (abbreviated CWC, iChemLabs, LLC) is a light-weight (~340 KB) JavaScript/HTML5 toolkit for chemical graphics, structure editing, interfaces, and informatics based on the proprietary ChemDoodle desktop software. The library uses <canvas> and WebGL technologies and other HTML5 features to provide solutions for creating chemistry-related applications for the web on desktop and mobile platforms. CWC can serve a broad range of scientific disciplines including crystallography, materials science, organic and inorganic chemistry, biochem- istry and chemical biology. CWC is freely available for in-house use and is open source (GPL v3) for all other uses. Keywords: ChemDoodle Web Components, Chemical graphics, Animations, Cheminformatics, HTML5, Canvas, JavaScript, WebGL, Structure editor, Structure query Introduction Mobile browsers did support HTML5, which opened How we communicate chemical information is increas- the door to web applications built with only HTML, ingly technology driven. Learning management systems, CSS and JavaScript (JS), such as the ChemDoodle Web virtual classrooms and MOOCs are a few examples where Components. chemistry educators need forward compatible tools for digital natives. Companies that implement emerg- Review ing web technologies can find efficiencies and benefit The ChemDoodle Web Components technology stack from competitive advantages. The first chemical graph- and features ics toolkit for the web, MDL Chime, was introduced in The ChemDoodle Web Components library, released in 1996 [1]. Based on the molecular visualization program 2009, is the first chemistry toolkit for structure viewing RasMol, Chime was developed as a plugin for Netscape and editing that is originally built using only web stand- and later for Internet Explorer and Firefox. -

CCP4 Molecular Graphics Documentation Contents Documentation On-Line Documentation Tutorials Ccp4mg Home Contents Quick Reference Print Version Contact References
CCP4 Molecular Graphics file:///E:/ccp4mg-win/help/index.html CCP4 Molecular Graphics Documentation Contents Documentation On-line Documentation Tutorials CCP4mg Home Contents Quick Reference Print version Contact References Documentation Contents General Graphics Window - mouse bindings and short cuts The Tools menu The View menu Projects - data organisation Installation of support programs Molecular models Reading coordinate files and atom typing The Coordinate Model Interface Atom selection Structure Analysis Surfaces and Electrostatic Potentials Symmetry Related Models Animations Other types of data Electron density maps Vectors Electron density in GAUSSIAN cube format Text and imported images Applications Superpose automatic protein superposition and interactive superposition of any structures Presentation Graphics Picture Wizard set up complex pictures quickly Movies Presentations Geometry Run Coot Other utilities Web tools History and Scripting Editing and Saving Files PDF Creator - PDF4Free v2.0 http://www.pdf4free.com 1 of 3 01/11/2007 18:42 CCP4 Molecular Graphics file:///E:/ccp4mg-win/help/index.html Lighting Saving the program status Picture Definition Files (also Object attributes and Introduction to Python) Making presentations Quick Reference See Display Table for general overview of the display table. For information on specific type of data click the appropriate data type in the File list below. File Tools View Applications Coordinate Files General tools General tools Movies Downloading Coordinate .. .. Superpose Files Read electron density Picture Save/restore view Lighting map/MTZ Wizard Read QM maps Save/restore status Transparency Geometry Add vectors/labels Picture definition files View from Rotate,translate,align Add image History and Scripting view Add legend Presentation/Notebook RocknRoll Add crystal List monomer definition Model definition file Preferences Open presentation Tutorials Output screen image Windows Bring a 'lost' window to the front. -

Coot: Model-Building Tools for Molecular Graphics Crystallography ISSN 0907-4449
research papers Acta Crystallographica Section D Biological Coot: model-building tools for molecular graphics Crystallography ISSN 0907-4449 Paul Emsley* and Kevin Cowtan CCP4mg is a project that aims to provide a general-purpose Received 26 February 2004 tool for structural biologists, providing tools for X-ray Accepted 4 August 2004 structure solution, structure comparison and analysis, and York Structural Biology Laboratory, University of York, Heslington, York YO10 5YW, England publication-quality graphics. The map-®tting tools are avail- able as a stand-alone package, distributed as `Coot'. Correspondence e-mail: [email protected] 1. Introduction Molecular graphics still plays an important role in the deter- mination of protein structures using X-ray crystallographic data, despite on-going efforts to automate model building. Functions such as side-chain placement, loop, ligand and fragment ®tting, structure comparison, analysis and validation are routinely performed using molecular graphics. Lower Ê resolution (dmin worse than 2.5 A) data in particular need interactive ®tting. The introduction of FRODO (Jones, 1978) and then O (Jones et al., 1991) to the ®eld of protein crystallography was in each case revolutionary, each in their time breaking new ground in demonstrating what was possible with the current hardware. These tools allowed protein crystallographers to enjoy what is widely held to be the most thrilling part of their work: giving birth (as it were) to a new protein structure. The CCP4 program suite (Collaborative Computational Project, Number 4, 1994) is an integrated collection of software for macromolecular crystallography, with a scope ranging from data processing to structure re®nement and validation. -
Instructions for PDB Downloading (From Either Website)
Molecular Visualization A BBSI Tutorial http://www.ccbb.pitt.edu/BBSI/index.htm By: Jeffry D. Madura Joshua A. Plumley Thomas J. Dick Table of Contents Instructions for PDB downloading..........................................................2 RasMol Tutorial ...........................................................................................3 VMD Tutorial ...............................................................................................5 CAChe Tutuorial..........................................................................................8 MOE Tutorial ............................................................................................. 10 Chimera Tutorial ....................................................................................... 13 Exercises .................................................................................................... 16 1 Instructions for PDB downloading (from either website) -Go to: - Go to: www.rcsb.org/pdb/ www.pdb.bu.edu/oca-bin/pdblite -Type in name of protein (examples at - Type in name of Protein / bottom of the page). Macromolecule ( examples at bottom of page). - Click on Name icon (first name in purple box). - Click on Retrieve Released Data Matching Your Query icon. -On the left side of the screen, click on Download/Display Structure - Highlight any of the molecule name and click on the View/ Analyze/ -Under Download the Structure File, Save Macro Molecule icon. right click on the X where the PDB(top) meets with none, under - Under the Data Retrieval section, -

USER MANUAL Version 4.5
GROMACS Groningen Machine for Chemical Simulations USER MANUAL Version 4.5 GROMACS USER MANUAL Version 4.5 Written by Emile Apol, Rossen Apostolov, Herman J.C. Berendsen, Aldert van Buuren, Par¨ Bjelkmar, Rudi van Drunen, Anton Feenstra, Gerrit Groenhof, Peter Kasson, Per Larsson, Peiter Meulenhoff, Teemu Murtola, Szilard´ Pall,´ Sander Pronk, Roland Schultz, Michael Shirts, Alfons Sijbers, Peter Tieleman Berk Hess, David van der Spoel, and Erik Lindahl. Additional contributions by Mark Abraham, Christoph Junghans, Carsten Kutzner, Justin A. Lemkul, Erik Marklund, Maarten Wolf. c 1991–2000: Department of Biophysical Chemistry, University of Groningen. Nijenborgh 4, 9747 AG Groningen, The Netherlands. c 2001–2010: The GROMACS development teams at the Royal Institute of Technology and Uppsala University, Sweden. More information can be found on our website: www.gromacs.org. iv Preface & Disclaimer This manual is not complete and has no pretention to be so due to lack of time of the contributors – our first priority is to improve the software. It is worked on continuously, which in some cases might mean the information is not entirely correct. Comments are welcome, please send them by e-mail to [email protected], or to one of the mailing lists (see www.gromacs.org). We try to release an updated version of the manual whenever we release a new version of the soft- ware, so in general it is a good idea to use a manual with the same major and minor release number as your GROMACS installation. Any revision numbers (like 3.1.1) are however independent, to make it possible to implement bug fixes and manual improvements if necessary. -

Architecture De Dataflow Pour Des Systèmes Modulaires Et Génériques De Simulation De Plante Christophe Pradal
Architecture de dataflow pour des systèmes modulaires et génériques de simulation de plante Christophe Pradal To cite this version: Christophe Pradal. Architecture de dataflow pour des systèmes modulaires et génériques desim- ulation de plante. Modélisation et simulation. Université Montpellier, 2019. Français. NNT : 2019MONTS034. tel-02879776 HAL Id: tel-02879776 https://tel.archives-ouvertes.fr/tel-02879776 Submitted on 24 Jun 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. THESE POUR OBTENIR LE GRADE DE DOCTEUR DE L’UNIVERSITE DE MONTPELLIER PAR LA VALIDATION DES ACQUIS DE L’EXPERIENCE Année universitaire 2018-2019 En Informatique École doctorale I2S – Information, Structures, Systèmes Unité de recherche UMR AGAP – Université de Montpellier SYNTHÈSE DES TRAVAUX DE RECHERCHE Architecture de Dataflow pour des systèmes modulaires et génériques de simulation de plante Présentée par Christophe PRADAL Le 17 juillet 2019 Référent : Christophe GODIN RAPPORT DE GESTION Devant le jury composé de Christophe GODIN, Directeur de recherche, Inria, Lyon -

Understanding Performance of Protein Structural Classifiers
Understanding Performance of Protein Structural Classifiers Alper Sarikaya∗ Danielle Albers† Michael Gleicher‡ University of Wisconsin – Madison University of Wisconsin – Madison University of Wisconsin – Madison ABSTRACT of proteins. The individual molecular detail view shows a surface- Many bioinformatics applications utilize machine learning tech- abstracted three-dimensional view of the protein, providing a scaf- niques to create models for predicting which parts of proteins will fold to display data mapped to the surface in a manner similar to bind to targets. Understanding the results of these protein surface canonical molecular graphics applications such as PyMol [3]. How- binding classifiers is challenging, as the individual answers are em- ever, we group spatially-congruent results on the molecular surface bedded spatially on the surface of the molecules, yet the perfor- to help collapse the data into a small set of regions that supports mance needs to be understood over an entire corpus of molecules. interactive and guided exploration. Our analytical approach is real- In this project, we introduce a multi-scale approach for assessing ized by a prototype that we have applied to various structural bind- the performance of these structural classifiers, providing coordi- ing classifiers. nated views for both corpus level overviews as well as spatially- 2 TASK ANALYSIS embedded results on the three-dimensional structures of proteins. Our problem domain focuses on proteins: large macro-molecules Keywords: J.3.1 [Computer Applications]: Life and Medical Sci- that are composed of 20 naturally-occurring amino acids (residues) ences — Biology and Genetics; chained together in sequence. The residues, in turn, fold over one another to form the conformation that governs the protein’s biolog- 1 INTRODUCTION ical and chemical function. -
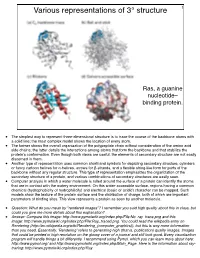
Various Representations of 3° Structure 1
Various representations of 3° structure 1 Ras, a guanine nucleotide– binding protein. • The simplest way to represent three-dimensional structure is to trace the course of the backbone atoms with a solid line; the most complex model shows the location of every atom. • The former shows the overall organization of the polypeptide chain without consideration of the amino acid side chains; the latter details the interactions among atoms that form the backbone and that stabilize the protein’s conformation. Even though both views are useful, the elements of secondary structure are not easily discerned in them. • Another type of representation uses common shorthand symbols for depicting secondary structure, cylinders or fancy cartoon helices for α-helices, arrows for β-strands, and a flexible string-like form for parts of the backbone without any regular structure. This type of representation emphasizes the organization of the secondary structure of a protein, and various combinations of secondary structures are easily seen. • Computer analysis in which a water molecule is rolled around the surface of a protein can identify the atoms that are in contact with the watery environment. On this water-accessible surface, regions having a common chemical (hydrophobicity or hydrophilicity) and electrical (basic or acidic) character can be mapped. Such models show the texture of the protein surface and the distribution of charge, both of which are important parameters of binding sites. This view represents a protein as seen by another molecule. • Question: What do you mean by "rendered images"? I remember you said high quality about this in class, but could you give me more details about this explanation? • Answer: Compare this image: http://www.pymolwiki.org/index.php/File:No_ray_trace.png and this image: http://www.pymolwiki.org/index.php/File:Ray_traced.png.