3D Printing of Molecular Models! 15!
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Pre-Activity Quiz Answer Key (Pdf)
Name: ____________________________________________ Date: ___________________ Class: ________________________ Pre-Activity Quiz Answer Key 1. Consider a molecule of carbon monoxide (C O): A. Do you think the electrons in the triple bond pull closer to the C atom or the O atom, or are they equally shared? Use the concept of electronegativity to explain your response. Answer: They are closer to the O atom. Using the electronegativity scale from a textbook, the result is a polar covalent bond shifted towards the O atom. B. Is the bond polar or non-polar? Polar 2. In today’s engineering challenge, you will sketch out Lewis dot diagrams for various molecules and polyatomic ions. Then you will construct each molecule using a molecular model kit. The kits contain three different representations: colored balls, short sticks and long flexible springs. A. Each colored ball corresponds to a different atom. How can you determine which color to use for each atom? Use the reference sheet that comes with the kit. B. For what bond type do you think the short sticks are used? Covalent bonds C. If you were to build a triple bond, what would you use to represent a triple bond and how many would you use? Use springs, three springs 3. You will become familiar with different geometries of simple molecules. A. Name the theory used to predict molecular shapes of these molecules? VSEPR theory Molecular Models and 3-D Printing Activity—Pre-Activity Quiz Answer Key 1 Name: ____________________________________________ Date: ___________________ Class: ________________________ B. What if a molecule contains a central atom bonded to two identical outer atoms with the central atom surrounded by a lone pair of electrons? Name the geometry of this molecule. -
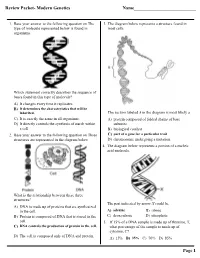
Review Packet- Modern Genetics Name___Page 1
Review Packet- Modern Genetics Name___________________________ 1. Base your answer to the following question on The 3. The diagram below represents a structure found in type of molecule represented below is found in most cells. organisms. Which statement correctly describes the sequence of bases found in this type of molecule? A) It changes every time it replicates. B) It determines the characteristics that will be inherited. The section labeled A in the diagram is most likely a C) It is exactly the same in all organisms. A) protein composed of folded chains of base D) It directly controls the synthesis of starch within subunits a cell. B) biological catalyst 2. Base your answer to the following question on Three C) part of a gene for a particular trait structures are represented in the diagram below. D) chromosome undergoing a mutation 4. The diagram below represents a portion of a nucleic acid molecule. What is the relationship between these three structures? The part indicated by arrow X could be A) DNA is made up of proteins that are synthesized in the cell. A) adenine B) ribose B) Protein is composed of DNA that is stored in the C) deoxyribose D) phosphate cell. 5. If 15% of a DNA sample is made up of thymine, T, C) DNA controls the production of protein in the cell. what percentage of the sample is made up of cytosine, C? D) The cell is composed only of DNA and protein. A) 15% B) 35% C) 70% D) 85% Page 1 6. Base your answer to the following question on Which 10.The diagram below represents genetic material. -

Application of X-Ray Diffraction Methods and Molecular Mechanics Simulations to Structure Determination and Cotton Fiber Analysis
University of New Orleans ScholarWorks@UNO University of New Orleans Theses and Dissertations Dissertations and Theses 12-19-2008 Application of X-ray Diffraction Methods and Molecular Mechanics Simulations to Structure Determination and Cotton Fiber Analysis Zakhia Moore University of New Orleans Follow this and additional works at: https://scholarworks.uno.edu/td Recommended Citation Moore, Zakhia, "Application of X-ray Diffraction Methods and Molecular Mechanics Simulations to Structure Determination and Cotton Fiber Analysis" (2008). University of New Orleans Theses and Dissertations. 888. https://scholarworks.uno.edu/td/888 This Dissertation is protected by copyright and/or related rights. It has been brought to you by ScholarWorks@UNO with permission from the rights-holder(s). You are free to use this Dissertation in any way that is permitted by the copyright and related rights legislation that applies to your use. For other uses you need to obtain permission from the rights-holder(s) directly, unless additional rights are indicated by a Creative Commons license in the record and/ or on the work itself. This Dissertation has been accepted for inclusion in University of New Orleans Theses and Dissertations by an authorized administrator of ScholarWorks@UNO. For more information, please contact [email protected]. Application of X-ray Diffraction Methods and Molecular Mechanics Simulations to Structure Determination and Cotton Fiber Analysis A Dissertation Submitted to the Graduate Faculty of the University of New Orleans in partial fulfillment of the Requirements for the Degree of Doctor of Philosophy in the Department of Chemistry by Zakhia Moore B.S., Xavier University, 2000 M.S., University of New Orleans, 2005 December, 2008 ACKNOWLEDGEMENTS Giving all glory to God for His many blessings, whom without I could not have completed this long journey. -

Journal of Molecular Graphics and Modelling 73 (2017) 179–190
Journal of Molecular Graphics and Modelling 73 (2017) 179–190 Contents lists available at ScienceDirect Journal of Molecular Graphics and Modelling journa l homepage: www.elsevier.com/locate/JMGM Combined Monte Carlo/torsion-angle molecular dynamics for ensemble modeling of proteins, nucleic acids and carbohydrates a b c d,1 Weihong Zhang , Steven C. Howell , David W. Wright , Andrew Heindel , e a, b, Xiangyun Qiu , Jianhan Chen ∗, Joseph E. Curtis ∗∗ a Kansas State University, Manhattan, KS, USA b NIST Center for Neutron Research, 100 Bureau Drive, Gaithersburg, MD, USA c Centre for Computational Science, Department of Chemistry, University College London, 10 Gordon St., London, UK d James Madison University, Department of Chemistry & Biochemistry, Harrisonburg, VA, USA e Department of Physics, The George Washington University, Washington, DC, USA a r t i c l e i n f o a b s t r a c t Article history: We describe a general method to use Monte Carlo simulation followed by torsion-angle molecular dynam- Received 13 October 2016 ics simulations to create ensembles of structures to model a wide variety of soft-matter biological systems. Received in revised form 19 January 2017 Our particular emphasis is focused on modeling low-resolution small-angle scattering and reflectivity Accepted 17 February 2017 structural data. We provide examples of this method applied to HIV-1 Gag protein and derived fragment Available online 23 February 2017 proteins, TraI protein, linear B-DNA, a nucleosome core particle, and a glycosylated monoclonal antibody. This procedure will enable a large community of researchers to model low-resolution experimental data MSC: with greater accuracy by using robust physics based simulation and sampling methods which are a 00-01 99-00 significant improvement over traditional methods used to interpret such data. -

IAPS Act 15-17 Section Preview of the Teachers Edition.Pdf
Section Preview of the Teacher’s Edition for The Chemistry of Materials, Issues and Physical Science, 2nd Edition Activities 15-17 Suggested student responses and answer keys have been blocked out so that web-savvy students do not find this page and have access to answers. To experience a complete activity please request a sample found in the footer at lab-aids.com Families of Elements 15 4 s 0 2 n - o t si o es 50 te s -minu N O I T I N A ACTIVITY OVERVIEW V E S T I G Students compare physical and chemical properties of 13 elements and sort them into groups based on common properties. They then compare their classifications with groups—or families—of elements as defined by scientists and displayed in the Periodic Table of the Elements. KEY CONCEPTS AND PROCESS SKILLS (with correlation to NSE 5–8 Content Standards) 1. Elements have characteristic physical and chemical properties. Substances often are placed in categories or groups if they react in similar ways; metals are an example of such a group. (PhysSci: 1) 2. Scientists classify elements into families, based on their properties. (PhysSci: 1) 3. Students develop descriptions based on evidence. (Inquiry: 1) KEY VOCABULARY atom atomic mass element family (of elements) metal Periodic Table of the Elements B-39 Activity 15 • Families of Elements MATERIALS AND ADVANCE PREPARATION For the teacher aluminum, iron, copper, and carbon samples from Activity 14, “Physical and Chemical Properties of Materials” 1 Transparency 15.1, “Information on Element Cards” 8 sets of 4 Element Family Cards -

Biological Sciences 1
Biological Sciences 1 BIOL UN3208 Introduction to Evolutionary Biology or EEEB UN2001 BIOLOGICAL SCIENCES Environmental Biology I: Elements to Organisms. Departmental Office: 600 Fairchild, 212-854-4581; Interested students should consult listings in other departments for [email protected]; [email protected] courses related to biology. For courses in environmental studies, see listings for Earth and environmental sciences or for ecology, evolution, Director of Undergraduate Studies, Undergraduate Programs and and environmental biology. For courses in human evolution, see listings Laboratories: for anthropology or for ecology, evolution, and environmental biology. Prof. Deborah Mowshowitz, 744D Mudd; 212-854-4497; For courses in the history of evolution, see listings for history and for [email protected] philosophy of science. For a list of courses in computational biology and genomics, visit http://systemsbiology.columbia.edu/courses. Biology Major and Concentration Advisers: For a list of current biology, biochemistry, biophysics, and neuroscience and behavior advisers, please visit http://biology.columbia.edu/ Advanced Placement programs/advisors Transfer Credit A-F: Prof. Alice Heicklen, 744B Mudd; [email protected] Transfer credits granted toward the degree are not automatically counted G-O: Prof. Mary Ann Price, 744A Mudd; [email protected] toward the major. The department determines which transfer credits P-Z: Prof. Tulle Hazelrigg, 753A Mudd, [email protected] can be counted toward the major. For most majors, at least four biology Backup Advisor: Prof. Deborah Mowshowitz, 744D Mudd; 212-854-4497; or biochemistry courses and at least 18 credits of the total (biology, [email protected] biochemistry, math, physics, and chemistry) must be taken at Columbia. -

CCP4 Molecular Graphics Documentation Contents Documentation On-Line Documentation Tutorials Ccp4mg Home Contents Quick Reference Print Version Contact References
CCP4 Molecular Graphics file:///E:/ccp4mg-win/help/index.html CCP4 Molecular Graphics Documentation Contents Documentation On-line Documentation Tutorials CCP4mg Home Contents Quick Reference Print version Contact References Documentation Contents General Graphics Window - mouse bindings and short cuts The Tools menu The View menu Projects - data organisation Installation of support programs Molecular models Reading coordinate files and atom typing The Coordinate Model Interface Atom selection Structure Analysis Surfaces and Electrostatic Potentials Symmetry Related Models Animations Other types of data Electron density maps Vectors Electron density in GAUSSIAN cube format Text and imported images Applications Superpose automatic protein superposition and interactive superposition of any structures Presentation Graphics Picture Wizard set up complex pictures quickly Movies Presentations Geometry Run Coot Other utilities Web tools History and Scripting Editing and Saving Files PDF Creator - PDF4Free v2.0 http://www.pdf4free.com 1 of 3 01/11/2007 18:42 CCP4 Molecular Graphics file:///E:/ccp4mg-win/help/index.html Lighting Saving the program status Picture Definition Files (also Object attributes and Introduction to Python) Making presentations Quick Reference See Display Table for general overview of the display table. For information on specific type of data click the appropriate data type in the File list below. File Tools View Applications Coordinate Files General tools General tools Movies Downloading Coordinate .. .. Superpose Files Read electron density Picture Save/restore view Lighting map/MTZ Wizard Read QM maps Save/restore status Transparency Geometry Add vectors/labels Picture definition files View from Rotate,translate,align Add image History and Scripting view Add legend Presentation/Notebook RocknRoll Add crystal List monomer definition Model definition file Preferences Open presentation Tutorials Output screen image Windows Bring a 'lost' window to the front. -

Coot: Model-Building Tools for Molecular Graphics Crystallography ISSN 0907-4449
research papers Acta Crystallographica Section D Biological Coot: model-building tools for molecular graphics Crystallography ISSN 0907-4449 Paul Emsley* and Kevin Cowtan CCP4mg is a project that aims to provide a general-purpose Received 26 February 2004 tool for structural biologists, providing tools for X-ray Accepted 4 August 2004 structure solution, structure comparison and analysis, and York Structural Biology Laboratory, University of York, Heslington, York YO10 5YW, England publication-quality graphics. The map-®tting tools are avail- able as a stand-alone package, distributed as `Coot'. Correspondence e-mail: [email protected] 1. Introduction Molecular graphics still plays an important role in the deter- mination of protein structures using X-ray crystallographic data, despite on-going efforts to automate model building. Functions such as side-chain placement, loop, ligand and fragment ®tting, structure comparison, analysis and validation are routinely performed using molecular graphics. Lower Ê resolution (dmin worse than 2.5 A) data in particular need interactive ®tting. The introduction of FRODO (Jones, 1978) and then O (Jones et al., 1991) to the ®eld of protein crystallography was in each case revolutionary, each in their time breaking new ground in demonstrating what was possible with the current hardware. These tools allowed protein crystallographers to enjoy what is widely held to be the most thrilling part of their work: giving birth (as it were) to a new protein structure. The CCP4 program suite (Collaborative Computational Project, Number 4, 1994) is an integrated collection of software for macromolecular crystallography, with a scope ranging from data processing to structure re®nement and validation. -

Molecular Model-Building by Computer
Molecular Model-building by Computer In which biochemists observe models of giant molecules as they are displayed on a screen by a computer and try to fold them into the shapes that they assume in nature by Cyrus Levinthal any problems of modern biology a large number of problems in biology computer to help gain such information are concerned with the detailed into which biologists have some insight about the structure of large biological M relation between biological but concerning which they cannot yet molecules. function and molecular structure. Some ask suitable questions in terms of molec- For the first half of this century the of the questions currently being asked ular structure. As they see such prob- metabolic and structural relations among will be completely answered only when lems more clearly, however, they in- the small molecules of the living cell one has an understanding of the struc- variably find an increasing need for were the principal concern of bio- ture of all the molecular components of structural information. In our laboratory chemists. The chemical reactions these a biological system and a knowledge of at the Massachusetts Institute of Tech- molecules undergo have been studied how they interact. There are, of course, nology we have recently started using a intensively. Such reactions are specifical- ly catalyzed by the large protein mole- cules called enzymes, many of which have now been purified and also studied. It is only within the past few years, however, that X-ray-diffraction techniques have made it possible to determine the molecular structure of such protein molecules. -

Architecture De Dataflow Pour Des Systèmes Modulaires Et Génériques De Simulation De Plante Christophe Pradal
Architecture de dataflow pour des systèmes modulaires et génériques de simulation de plante Christophe Pradal To cite this version: Christophe Pradal. Architecture de dataflow pour des systèmes modulaires et génériques desim- ulation de plante. Modélisation et simulation. Université Montpellier, 2019. Français. NNT : 2019MONTS034. tel-02879776 HAL Id: tel-02879776 https://tel.archives-ouvertes.fr/tel-02879776 Submitted on 24 Jun 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. THESE POUR OBTENIR LE GRADE DE DOCTEUR DE L’UNIVERSITE DE MONTPELLIER PAR LA VALIDATION DES ACQUIS DE L’EXPERIENCE Année universitaire 2018-2019 En Informatique École doctorale I2S – Information, Structures, Systèmes Unité de recherche UMR AGAP – Université de Montpellier SYNTHÈSE DES TRAVAUX DE RECHERCHE Architecture de Dataflow pour des systèmes modulaires et génériques de simulation de plante Présentée par Christophe PRADAL Le 17 juillet 2019 Référent : Christophe GODIN RAPPORT DE GESTION Devant le jury composé de Christophe GODIN, Directeur de recherche, Inria, Lyon -

Understanding Performance of Protein Structural Classifiers
Understanding Performance of Protein Structural Classifiers Alper Sarikaya∗ Danielle Albers† Michael Gleicher‡ University of Wisconsin – Madison University of Wisconsin – Madison University of Wisconsin – Madison ABSTRACT of proteins. The individual molecular detail view shows a surface- Many bioinformatics applications utilize machine learning tech- abstracted three-dimensional view of the protein, providing a scaf- niques to create models for predicting which parts of proteins will fold to display data mapped to the surface in a manner similar to bind to targets. Understanding the results of these protein surface canonical molecular graphics applications such as PyMol [3]. How- binding classifiers is challenging, as the individual answers are em- ever, we group spatially-congruent results on the molecular surface bedded spatially on the surface of the molecules, yet the perfor- to help collapse the data into a small set of regions that supports mance needs to be understood over an entire corpus of molecules. interactive and guided exploration. Our analytical approach is real- In this project, we introduce a multi-scale approach for assessing ized by a prototype that we have applied to various structural bind- the performance of these structural classifiers, providing coordi- ing classifiers. nated views for both corpus level overviews as well as spatially- 2 TASK ANALYSIS embedded results on the three-dimensional structures of proteins. Our problem domain focuses on proteins: large macro-molecules Keywords: J.3.1 [Computer Applications]: Life and Medical Sci- that are composed of 20 naturally-occurring amino acids (residues) ences — Biology and Genetics; chained together in sequence. The residues, in turn, fold over one another to form the conformation that governs the protein’s biolog- 1 INTRODUCTION ical and chemical function. -

CORINA Classic Program Manual
CORINA Classic Generation of Three-Dimensional Molecular Models Version 4.4.0 Program Description Molecular Networks GmbH September 2021 www.mn-am.com Molecular Networks GmbH Altamira LLC Neumeyerstraße 28 470 W Broad St, Unit #5007 90411 Nuremberg Columbus, OH 43215 Germany USA mn-am.com This document is copyright © 1998-2021 by Molecular Networks GmbH Computerchemie and Altamira LLC. All rights reserved. Except as permitted under the terms of the Software Licensing Agreement of Molecular Networks GmbH Computerchemie, no part of this publication may be reproduced or distributed in any form or by any means or stored in a database retrieval system without the prior written permission of Molecular Networks GmbH or Altamira LLC. The software described in this document is furnished under a license and may be used and copied only in accordance with the terms of such license. (Document version: 4.4.0-2021-09-30) Content Content 1 Introducing CORINA Classic 1 1.1 Objective of CORINA Classic 1 1.2 CORINA Classic in Brief 1 2 Release Notes 3 2.1 CORINA (Full Version) 3 2.2 CORINA_F (Restricted FlexX Interface Version) 25 3 Getting Started with CORINA Classic 26 4 Using CORINA Classic 29 4.1 Synopsis 29 4.2 Options 29 5 Use Cases of CORINA Classic 60 6 Supported File Formats and Interfaces 64 6.1 V2000 Structure Data File (SD) and Reaction Data File (RD) 64 6.2 V3000 Structure Data File (SD) and Reaction Data File (RD) 67 6.3 SMILES Linear Notation 67 6.4 InChI file format 69 6.5 SYBYL File Formats 70 6.6 Brookhaven Protein Data Bank Format (PDB)