View of the Entific) Were Incubated in 1 × T4 PNK Buffer (Thermo Pomago Alignment in the Supplemental Material
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diversity of Understudied Archaeal and Bacterial Populations of Yellowstone National Park: from Genes to Genomes Daniel Colman
University of New Mexico UNM Digital Repository Biology ETDs Electronic Theses and Dissertations 7-1-2015 Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes Daniel Colman Follow this and additional works at: https://digitalrepository.unm.edu/biol_etds Recommended Citation Colman, Daniel. "Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes." (2015). https://digitalrepository.unm.edu/biol_etds/18 This Dissertation is brought to you for free and open access by the Electronic Theses and Dissertations at UNM Digital Repository. It has been accepted for inclusion in Biology ETDs by an authorized administrator of UNM Digital Repository. For more information, please contact [email protected]. Daniel Robert Colman Candidate Biology Department This dissertation is approved, and it is acceptable in quality and form for publication: Approved by the Dissertation Committee: Cristina Takacs-Vesbach , Chairperson Robert Sinsabaugh Laura Crossey Diana Northup i Diversity of understudied archaeal and bacterial populations from Yellowstone National Park: from genes to genomes by Daniel Robert Colman B.S. Biology, University of New Mexico, 2009 DISSERTATION Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biology The University of New Mexico Albuquerque, New Mexico July 2015 ii DEDICATION I would like to dedicate this dissertation to my late grandfather, Kenneth Leo Colman, associate professor of Animal Science in the Wool laboratory at Montana State University, who even very near the end of his earthly tenure, thought it pertinent to quiz my knowledge of oxidized nitrogen compounds. He was a man of great curiosity about the natural world, and to whom I owe an acknowledgement for his legacy of intellectual (and actual) wanderlust. -
A Deep Learning Approach to Pattern Recognition for Short DNA Sequences”
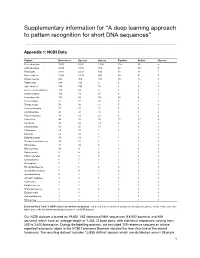
Supplementary information for “A deep learning approach to pattern recognition for short DNA sequences” Appendix 1: NCBI Data Phylum References Species Genera Families Orders Classes Proteobacteria 7,053 5,061 1,106 158 55 9 Actinobacteria 4,768 3,313 383 68 29 6 Firmicutes 3,814 2,531 499 56 13 7 Bacteroidetes 1,934 1,525 360 39 8 7 Euryarchaeota 834 450 100 28 13 8 Tenericutes 266 195 8 5 4 1 Spirochaetes 146 100 16 6 4 1 Deinococcus-Thermus 118 99 9 3 2 1 Crenarchaeota 113 61 27 8 5 1 Cyanobacteria 113 88 59 30 8 2 Fusobacteria 75 37 10 2 1 1 Thermotogae 70 48 13 5 4 1 Verrucomicrobia 57 53 22 7 4 3 Acidobacteria 45 41 19 5 5 4 Planctomycetes 44 31 23 5 3 2 Chloroflexi 44 35 26 15 12 8 Aquificae 43 32 14 4 2 1 Synergistetes 31 25 15 1 1 1 Chlamydiae 28 18 7 5 2 1 Chlorobi 21 16 5 1 1 1 Deferribacteres 15 11 7 1 1 1 Thermodesulfobacteria 14 12 5 1 1 1 Nitrospirae 11 10 3 1 1 1 Fibrobacteres 10 4 3 3 3 3 Balneolaeota 9 9 4 1 1 1 Chrysiogenetes 6 4 3 1 1 1 Lentisphaerae 6 5 3 3 3 2 Dictyoglomi 5 2 1 1 1 1 Rhodothermaeota 5 5 3 2 1 1 Gemmatimonadetes 5 4 3 2 2 2 Ignavibacteriae 4 2 2 2 1 1 Armatimonadetes 4 3 3 3 3 3 Caldiserica 3 1 1 1 1 1 Calditrichaeota 2 2 1 1 1 1 Thaumarchaeota 2 2 2 2 2 2 Elusimicrobia 2 1 1 1 1 1 Kiritimatiellaeota 1 1 1 1 1 1 Nitrospinae 1 1 1 1 1 1 Extended Data Table 1: NCBI dataset breakdown by phylum. -

Thermophilic Lithotrophy and Phototrophy in an Intertidal, Iron-Rich, Geothermal Spring 2 3 Lewis M
bioRxiv preprint doi: https://doi.org/10.1101/428698; this version posted September 27, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Thermophilic Lithotrophy and Phototrophy in an Intertidal, Iron-rich, Geothermal Spring 2 3 Lewis M. Ward1,2,3*, Airi Idei4, Mayuko Nakagawa2,5, Yuichiro Ueno2,5,6, Woodward W. 4 Fischer3, Shawn E. McGlynn2* 5 6 1. Department of Earth and Planetary Sciences, Harvard University, Cambridge, MA 02138 USA 7 2. Earth-Life Science Institute, Tokyo Institute of Technology, Meguro, Tokyo, 152-8550, Japan 8 3. Division of Geological and Planetary Sciences, California Institute of Technology, Pasadena, CA 9 91125 USA 10 4. Department of Biological Sciences, Tokyo Metropolitan University, Hachioji, Tokyo 192-0397, 11 Japan 12 5. Department of Earth and Planetary Sciences, Tokyo Institute of Technology, Meguro, Tokyo, 13 152-8551, Japan 14 6. Department of Subsurface Geobiological Analysis and Research, Japan Agency for Marine-Earth 15 Science and Technology, Natsushima-cho, Yokosuka 237-0061, Japan 16 Correspondence: [email protected] or [email protected] 17 18 Abstract 19 Hydrothermal systems, including terrestrial hot springs, contain diverse and systematic 20 arrays of geochemical conditions that vary over short spatial scales due to progressive interaction 21 between the reducing hydrothermal fluids, the oxygenated atmosphere, and in some cases 22 seawater. At Jinata Onsen, on Shikinejima Island, Japan, an intertidal, anoxic, iron- and 23 hydrogen-rich hot spring mixes with the oxygenated atmosphere and sulfate-rich seawater over 24 short spatial scales, creating an enormous range of redox environments over a distance ~10 m. -

7. References
University of Akureyri Department of Natural Resource Science 7. References Ammann, E. C., Reed, L. L., & Durichek, J. J. (1968). Gas consumptions and Growth Rate of Hydrogenomonas eutropha in Continuous Culture. Applied Microbiology , 16, (6), 822-826. Aguiar, P., Beveridge, T. J., & Reysenbach, A.-L. (2004). Sulfurihydrogenibium azorense, sp. nov., a thermophilic hydrogen oxidizing microaerophile from terrestrial hot springs in the Azores. International Journal of Systematic and Evolutionary Microbiology , 54, 33-39. Altschul, S., Gish, W., Miller, W., Myers, E., & Lipman, D. (1990). "Basic local alignment search tool.". J. Mol. Biol. , 215:403-410. Amend, J., & Shock, E. (2001). Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiology Reviews , 25, 175-243. Aragno, M. (1978). Enrichment, isolation and preliminary characterization of a thermophilic, endospore-forming hydrogen bacterium. FEMS Micobiol. Lett. , 3: 13-15. Aragno, M. (1992). The Thermophilic, Aerobic, Hydrogen-Oxidizing (Knallgas) Bacteria. In A. Balows, H. Trüper, M. Dworkin, W. Harder, & K. Schleifer, The Prokaryotes, a handbook on biology of bacteria. 2nd ed. vol. 4 (pp. 3917-3933.). New York: Springer Verlag. Aragno, M., & Schlegel, H. G. (1992). The mesophilic Hydrogen-Oxidizing (Knallgas) Bacteria. In A. Balows, H. Truper, M. Dworkin, W. Harder, & K.-H. Schleifer, The Prokaryotes 2nd. ed. (pp. 344-384). New York: Springer. Ármannson, H. (2002, May 30-31). Erindi á ráðstefnu um málefni veitufyrirtækja . Grænt bókhald í jarðhita- samanburður á útblæstri við aðra orkugjafa . Akureyri, Iceland: Samorka. Bae, S., Kwak, K., Kim, S., Chung, S., & Igarashi, Y. (2001). Isolation and Characterization of CO2-Fixing Hydrogen -Oxidizing Marine 109 University of Akureyri Department of Natural Resource Science Bacteria. -

Characterization of the Uncommon Enzymes from (2004)
OPEN Mannosylglucosylglycerate biosynthesis SUBJECT AREAS: in the deep-branching phylum WATER MICROBIOLOGY MARINE MICROBIOLOGY Planctomycetes: characterization of the HOMEOSTASIS MULTIENZYME COMPLEXES uncommon enzymes from Rhodopirellula Received baltica 13 March 2013 Sofia Cunha1, Ana Filipa d’Avo´1, Ana Mingote2, Pedro Lamosa3, Milton S. da Costa1,4 & Joana Costa1,4 Accepted 23 July 2013 1Center for Neuroscience and Cell Biology, University of Coimbra, 3004-517 Coimbra, Portugal, 2Instituto de Tecnologia Quı´mica Published e Biolo´gica, Universidade Nova de Lisboa, 2780-157 Oeiras, Portugal, 3Centro de Ressonaˆncia Magne´tica Anto´nio Xavier, 7 August 2013 Instituto de Tecnologia Quı´mica e Biolo´gica, Universidade Nova de Lisboa, 2781-901 Oeiras, Portugal, 4Department of Life Sciences, University of Coimbra, Apartado 3046, 3001-401 Coimbra, Portugal. Correspondence and The biosynthetic pathway for the rare compatible solute mannosylglucosylglycerate (MGG) accumulated by requests for materials Rhodopirellula baltica, a marine member of the phylum Planctomycetes, has been elucidated. Like one of the should be addressed to pathways used in the thermophilic bacterium Petrotoga mobilis, it has genes coding for J.C. ([email protected].) glucosyl-3-phosphoglycerate synthase (GpgS) and mannosylglucosyl-3-phosphoglycerate (MGPG) synthase (MggA). However, unlike Ptg. mobilis, the mesophilic R. baltica uses a novel and very specific MGPG phosphatase (MggB). It also lacks a key enzyme of the alternative pathway in Ptg. mobilis – the mannosylglucosylglycerate synthase (MggS) that catalyses the condensation of glucosylglycerate with GDP-mannose to produce MGG. The R. baltica enzymes GpgS, MggA, and MggB were expressed in E. coli and characterized in terms of kinetic parameters, substrate specificity, temperature and pH dependence. -

And Thermo-Adaptation in Hyperthermophilic Archaea: Identification of Compatible Solutes, Accumulation Profiles, and Biosynthetic Routes in Archaeoglobus Spp
Universidade Nova de Lisboa Osmo- andInstituto thermo de Tecnologia-adaptation Química e Biológica in hyperthermophilic Archaea: Subtitle Subtitle Luís Pedro Gafeira Gonçalves Osmo- and thermo-adaptation in hyperthermophilic Archaea: identification of compatible solutes, accumulation profiles, and biosynthetic routes in Archaeoglobus spp. OH OH OH CDP c c c - CMP O O - PPi O3P P CTP O O O OH OH OH OH OH OH O- C C C O P O O P i Dissertation presented to obtain the Ph.D degree in BiochemistryO O- Instituto de Tecnologia Química e Biológica | Universidade Nova de LisboaP OH O O OH OH OH Oeiras, Luís Pedro Gafeira Gonçalves January, 2008 2008 Universidade Nova de Lisboa Instituto de Tecnologia Química e Biológica Osmo- and thermo-adaptation in hyperthermophilic Archaea: identification of compatible solutes, accumulation profiles, and biosynthetic routes in Archaeoglobus spp. This dissertation was presented to obtain a Ph. D. degree in Biochemistry at the Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa. By Luís Pedro Gafeira Gonçalves Supervised by Prof. Dr. Helena Santos Oeiras, January, 2008 Apoio financeiro da Fundação para a Ciência e Tecnologia (POCI 2010 – Formação Avançada para a Ciência – Medida IV.3) e FSE no âmbito do Quadro Comunitário de apoio, Bolsa de Doutoramento com a referência SFRH / BD / 5076 / 2001. ii ACKNOWNLEDGMENTS The work presented in this thesis, would not have been possible without the help, in terms of time and knowledge, of many people, to whom I am extremely grateful. Firstly and mostly, I need to thank my supervisor, Prof. Helena Santos, for her way of thinking science, her knowledge, her rigorous criticism, and her commitment to science. -

BEING Aquifex Aeolicus: UNTANGLING a HYPERTHERMOPHILE‘S CHECKERED PAST
BEING Aquifex aeolicus: UNTANGLING A HYPERTHERMOPHILE‘S CHECKERED PAST by Robert J.M. Eveleigh Submitted in partial fulfillment of the requirements for the degree of Master of Science at Dalhousie University Halifax, Nova Scotia December 2011 © Copyright by Robert J.M. Eveleigh, 2011 DALHOUSIE UNIVERSITY DEPARTMENT OF COMPUTATIONAL BIOLOGY AND BIOINFORMATICS The undersigned hereby certify that they have read and recommend to the Faculty of Graduate Studies for acceptance a thesis entitled ―BEING Aquifex aeolicus: UNTANGLING A HYPERTHERMOPHILE‘S CHECKERED PAST‖ by Robert J.M. Eveleigh in partial fulfillment of the requirements for the degree of Master of Science. Dated: December 13, 2011 Co-Supervisors: _________________________________ _________________________________ Readers: _________________________________ ii DALHOUSIE UNIVERSITY DATE: December 13, 2011 AUTHOR: Robert J.M. Eveleigh TITLE: BEING Aquifex aeolicus: UNTANGLING A HYPERTHERMOPHILE‘S CHECKERED PAST DEPARTMENT OR SCHOOL: Department of Computational Biology and Bioinformatics DEGREE: MSc CONVOCATION: May YEAR: 2012 Permission is herewith granted to Dalhousie University to circulate and to have copied for non-commercial purposes, at its discretion, the above title upon the request of individuals or institutions. I understand that my thesis will be electronically available to the public. The author reserves other publication rights, and neither the thesis nor extensive extracts from it may be printed or otherwise reproduced without the author‘s written permission. The author attests that permission has been obtained for the use of any copyrighted material appearing in the thesis (other than the brief excerpts requiring only proper acknowledgement in scholarly writing), and that all such use is clearly acknowledged. _______________________________ Signature of Author iii TABLE OF CONTENTS List of Tables .................................................................................................................... -

Lewis M. Ward *, Airi Idei , Mayuko Nakagawa , Yuichiro
bioRxiv preprint doi: https://doi.org/10.1101/428698; this version posted January 27, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Geochemical and metagenomic characterization of Jinata Onsen, a Proterozoic-analog hot spring, 2 reveals novel microbial diversity including iron-tolerant phototrophs and thermophilic lithotrophs 3 4 Lewis M. Ward1,2,3*, Airi Idei4, Mayuko Nakagawa2,5, Yuichiro Ueno2,5,6, Woodward W. 5 Fischer3, Shawn E. McGlynn2* 6 7 1. Department of Earth and Planetary Sciences, Harvard University, Cambridge, MA 02138 USA 8 2. Earth-Life Science Institute, Tokyo Institute of Technology, Meguro, Tokyo, 152-8550, Japan 9 3. Division of Geological and Planetary Sciences, California Institute of Technology, Pasadena, CA 10 91125 USA 11 4. Department of Biological Sciences, Tokyo Metropolitan University, Hachioji, Tokyo 192-0397, 12 Japan 13 5. Department of Earth and Planetary Sciences, Tokyo Institute of Technology, Meguro, Tokyo, 14 152-8551, Japan 15 6. Department of Subsurface Geobiological Analysis and Research, Japan Agency for Marine-Earth 16 Science and Technology, Natsushima-cho, Yokosuka 237-0061, Japan 17 Correspondence: [email protected] or [email protected] 18 19 Abstract 20 Hydrothermal systems, including terrestrial hot springs, contain diverse geochemical 21 conditions that vary over short spatial scales due to progressive interaction between the reducing 22 hydrothermal fluids, the oxygenated atmosphere, and in some cases seawater. At Jinata Onsen, 23 on Shikinejima Island, Japan, an intertidal, anoxic, iron-rich hot spring mixes with the 24 oxygenated atmosphere and seawater over short spatial scales, creating a diversity of chemical 25 potentials and redox pairs over a distance ~10 m. -

A Method for Achieving Complete Microbial Genomes and Improving Bins from Metagenomics Data
bioRxiv preprint doi: https://doi.org/10.1101/2020.03.05.979740; this version posted July 18, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. 1 A method for achieving complete microbial genomes and improving 2 bins from metagenomics data 3 4 Authors: 5 Lauren M. Lui1, Torben N. Nielsen1, Adam P. Arkin1,2,3* 6 7 Affiliations: 8 1Environmental Genomics and Systems Biology Division, Lawrence Berkeley National 9 Laboratory, Berkeley, CA, USA. 10 2Department of Bioengineering, University of California, Berkeley, CA, USA 11 3Innovative Genomics Institute, Berkeley, CA, USA 12 13 *Correspondence: [email protected] 14 15 16 17 18 19 20 21 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.03.05.979740; this version posted July 18, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. 22 Abstract 23 Metagenomics facilitates the study of the genetic information from uncultured microbes 24 and complex microbial communities. Assembling complete microbial genomes (i.e., 25 circular with no misassemblies) from metagenomics data is difficult because most 26 samples have high organismal complexity and strain diversity. Less than 100 27 circularized bacterial and archaeal genomes have been assembled from metagenomics 28 data despite the thousands of datasets that are available. -

Hydrogenobacter Thermophilus Type Strain (TK-6T)
Lawrence Berkeley National Laboratory Recent Work Title Complete genome sequence of Hydrogenobacter thermophilus type strain (TK-6). Permalink https://escholarship.org/uc/item/9071q43r Journal Standards in genomic sciences, 4(2) ISSN 1944-3277 Authors Zeytun, Ahmet Sikorski, Johannes Nolan, Matt et al. Publication Date 2011-04-01 DOI 10.4056/sigs.1463589 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Standards in Genomic Sciences (2011) 4:131-143 DOI:10.4056/sigs.1463589 Complete genome sequence of Hydrogenobacter thermophilus type strain (TK-6T) Ahmet Zeytun1,3, Johannes Sikorski2, Matt Nolan1, Alla Lapidus1, Susan Lucas1, James Han1, Hope Tice1, Jan-Fang Cheng1, Roxanne Tapia1,3, Lynne Goodwin1,3, Sam Pitluck1, Konstantinos Liolios1, Natalia Ivanova1, Konstantinos Mavromatis1, Natalia Mikhailova1, Galina Ovchinnikova1, Amrita Pati1, Amy Chen4, Krishna Palaniappan4, Olivier D. Ngatchou-Djao5, Miriam Land1,6, Loren Hauser1,6, Cynthia D. Jeffries1,6, Cliff Han1,3, John C. Detter1,3, Susanne Übler7, Manfred Rohde5, Brian J. Tindall2, Markus Göker2, Reinhard Wirth7, Tanja Woyke1, James Bristow1, Jonathan A. Eisen1,8, Victor Markowitz4, Philip Hugenholtz1,9, Hans-Peter Klenk2, and Nikos C. Kyrpides1* 1 DOE Joint Genome Institute, Walnut Creek, California, USA 2 DSMZ - German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig, Germany 3 Los Alamos National Laboratory, Bioscience Division, Los Alamos, New Mexico, USA 4 Biological Data Management and Technology Center, Lawrence Berkeley National Laboratory, Berkeley, California, USA 5 HZI – Helmholtz Centre for Infection Research, Braunschweig, Germany 6 Oak Ridge National Laboratory, Oak Ridge, Tennessee, USA 7 Archaea Centre - University of Regensburg, Regensburg, Germany 8 University of California Davis Genome Center, Davis, California, USA 9 Australian Centre for Ecogenomics, School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, Australia *Corresponding author: Nikos C. -

Thermovibrio Ammonificans HB-1T
Standards in Genomic Sciences (2012) 7:82-90 DOI:10.4056/sigs.2856770 Complete genome sequence of Thermovibrio ammonificans HB-1T, a thermophilic, chemolithoautotrophic bacterium isolated from a deep-sea hydrothermal vent Donato Giovannelli1,2,3, Jessica Ricci1,2, Ileana Pérez-Rodríguez1,2, Michael Hügler4, Charles O’Brien1, , Ramaydalis Keddis1,2, Ashley Grosche1,2, Lynne Goodwin6, David Bruce5, Karen W. Davenport6, Chris Detter6, James Han5, Shunsheng Han6, Natalia Ivanova5, Miriam L. Land7, Natalia Mikhailova5, Matt Nolan5, Sam Pitluck5, Roxanne Tapia6, Tanja Woyke5 and Costantino Vetriani1,2 1Department of Biochemistry and Microbiology, Rutgers University, New Brunswick, NJ, USA 2Institute of Marine and Coastal Sciences, Rutgers University, New Brunswick, NJ, USA 3Institute for Marine Science - ISMAR, National Research Council of Italy - CNR, Ancona, Italy 4Microbiology Department, Water Technology Center, Karlsruhe - Germany 5Joint Genome Institute, Walnut Creek, CA, USA 6Los Alamos National Laboratory, Bioscience Division, Los Alamos, NM, USA 7Oak Ridge National Laboratory, Oak Ridge, TN, USA Corresponding author: Costantino Vetriani ([email protected]) Keywords: Aquificae, Desulfurobacteriaceae, thermophilic, anaerobic, chemolithoautotrophic, hydrothermal vent T Thermovibrio ammonificans type strain HB-1 is a thermophilic (Topt: 75°C), strictly anaero- bic, chemolithoautotrophic bacterium that was isolated from an active, high temperature deep-sea hydrothermal vent on the East Pacific Rise. This organism grows on mineral salts - 0 medium in the presence of CO2/H2, using NO3 or S as electron acceptors, which are re- duced to ammonium or hydrogen sulfide, respectively. T. ammonificans is one of only three species within the genus Thermovibrio, a member of the family Desulfurobacteriaceae, and it forms a deep branch within the phylum Aquificae. -

Evidence for Massive Gene Exchange Between Archaeal and Bacterial
MEETING REPORT releasing Sir proteins from the be silenced by the Sir protein com- 3 Loo, S. and Rine, J. (1995) Annu. Ku70p–Ku80p telomerase complex plex. In summary, the importance of Rev. Cell Dev. Biol. 11, (David Shore, Univ. of Geneva, chromatin structure was evident in 519–548 Switzerland). Cdc13p protein binds all sessions. Yeast origins, cen- 4 Smith, J.S. and Boeke, J.D. (1997) single-stranded DNA at the tromeres and telomeres bind elegant Genes Dev. 11, 241–254 Ku70p–Ku80p telomerase complex multiprotein complexes that act as 5 Weaver, D.T. (1995) Trends Genet. (Vicki Lundblad, Baylor, USA). regulatory machines to change 11, 388–392 Nuclear organization of telomeres is chromatin structure and to allow important with telomeres located at important cellular processes to occur. the nuclear periphery (Sussan Robert A. Sclafani [email protected] Gasser, ISREC, Switzerland). Target- Further reading ting DNA to the periphery using a 1 Dutta, A. and Bell, S.P. (1997) ER–Golgi anchoring signal can pro- Annu. Rev. Cell Dev. Biol. 13, Department of Biochemistry and Molecular duce silencing (Rolf Sternglanz, 293–332 Genetics, University of Colorado Health SUNY, USA). Hence, any gene 2 Pluta, A.F. et al. (1995) Science 270, Sciences Center, 4200 E. Ninth Avenue, brought to the nuclear periphery will 1591–1594 Denver, CO 80262, USA. LETTER reasoned that a detailed comparison Evidence for massive gene exchange of the Aquifex and archaeal between archaeal and bacterial genomes could reveal genome-scale adaptations for thermophily. hyperthermophiles The protein sequences encoded in all complete bacterial genomes were compared with the non- redundant protein sequence Sequencing of multiple complete exceptional among bacteria in that it database using the gapped BLAST genomes of bacteria and archaea occupies the hyperthermophilic program7, and a phylogenetic makes it possible to perform niche otherwise dominated by breakdown was automatically systematic, genome-scale archaea2.