Uva-DARE (Digital Academic Repository)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Number 2 February 2014
Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL INIST -CNRS Volume 18 - Number 2 February 2014 The PDF version of the Atlas of Genetics and Cytogenetics in Oncology and Haematology is a reissue of the original articles published in collaboration with the Institute for Scientific and Technical Information (INstitut de l’Information Scientifique et Technique - INIST) of the French National Center for Scientific Research (CNRS) on its electronic publishing platform I-Revues. Online and PDF versions of the Atlas of Genetics and Cytogenetics in Oncology and Haematology are hosted by INIST-CNRS. Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL INIST -CNRS Scope The Atlas of Genetics and Cytogenetics in Oncology and Haematology is a peer reviewed on-line journal in open access, devoted to genes, cytogenetics, and clinical entities in cancer, and cancer-prone diseases. It presents structured review articles (“cards”) on genes, leukaemias, solid tumours, cancer-prone diseases, and also more traditional review articles (“deep insights”) on the above subjects and on surrounding topics. It also present case reports in hematology and educational items in the various related topics for students in Medicine and in Sciences. Editorial correspondance Jean-Loup Huret Genetics, Department of Medical Information, University Hospital F-86021 Poitiers, France tel +33 5 49 44 45 46 or +33 5 49 45 47 67 [email protected] or [email protected] Staff Mohammad Ahmad, Mélanie Arsaban, Marie-Christine Jacquemot-Perbal, Vanessa Le Berre, Anne Malo, Carol Moreau, Catherine Morel-Pair, Laurent Rassinoux, Alain Zasadzinski. Philippe Dessen is the Database Director, and Alain Bernheim the Chairman of the on-line version (Gustave Roussy Institute – Villejuif – France). -

List of Genes Associated with Sudden Cardiac Death (Scdgseta) Gene
List of genes associated with sudden cardiac death (SCDgseta) mRNA expression in normal human heart Entrez_I Gene symbol Gene name Uniprot ID Uniprot name fromb D GTEx BioGPS SAGE c d e ATP-binding cassette subfamily B ABCB1 P08183 MDR1_HUMAN 5243 √ √ member 1 ATP-binding cassette subfamily C ABCC9 O60706 ABCC9_HUMAN 10060 √ √ member 9 ACE Angiotensin I–converting enzyme P12821 ACE_HUMAN 1636 √ √ ACE2 Angiotensin I–converting enzyme 2 Q9BYF1 ACE2_HUMAN 59272 √ √ Acetylcholinesterase (Cartwright ACHE P22303 ACES_HUMAN 43 √ √ blood group) ACTC1 Actin, alpha, cardiac muscle 1 P68032 ACTC_HUMAN 70 √ √ ACTN2 Actinin alpha 2 P35609 ACTN2_HUMAN 88 √ √ √ ACTN4 Actinin alpha 4 O43707 ACTN4_HUMAN 81 √ √ √ ADRA2B Adrenoceptor alpha 2B P18089 ADA2B_HUMAN 151 √ √ AGT Angiotensinogen P01019 ANGT_HUMAN 183 √ √ √ AGTR1 Angiotensin II receptor type 1 P30556 AGTR1_HUMAN 185 √ √ AGTR2 Angiotensin II receptor type 2 P50052 AGTR2_HUMAN 186 √ √ AKAP9 A-kinase anchoring protein 9 Q99996 AKAP9_HUMAN 10142 √ √ √ ANK2/ANKB/ANKYRI Ankyrin 2 Q01484 ANK2_HUMAN 287 √ √ √ N B ANKRD1 Ankyrin repeat domain 1 Q15327 ANKR1_HUMAN 27063 √ √ √ ANKRD9 Ankyrin repeat domain 9 Q96BM1 ANKR9_HUMAN 122416 √ √ ARHGAP24 Rho GTPase–activating protein 24 Q8N264 RHG24_HUMAN 83478 √ √ ATPase Na+/K+–transporting ATP1B1 P05026 AT1B1_HUMAN 481 √ √ √ subunit beta 1 ATPase sarcoplasmic/endoplasmic ATP2A2 P16615 AT2A2_HUMAN 488 √ √ √ reticulum Ca2+ transporting 2 AZIN1 Antizyme inhibitor 1 O14977 AZIN1_HUMAN 51582 √ √ √ UDP-GlcNAc: betaGal B3GNT7 beta-1,3-N-acetylglucosaminyltransfe Q8NFL0 -

Investigation of Candidate Genes and Mechanisms Underlying Obesity
Prashanth et al. BMC Endocrine Disorders (2021) 21:80 https://doi.org/10.1186/s12902-021-00718-5 RESEARCH ARTICLE Open Access Investigation of candidate genes and mechanisms underlying obesity associated type 2 diabetes mellitus using bioinformatics analysis and screening of small drug molecules G. Prashanth1 , Basavaraj Vastrad2 , Anandkumar Tengli3 , Chanabasayya Vastrad4* and Iranna Kotturshetti5 Abstract Background: Obesity associated type 2 diabetes mellitus is a metabolic disorder ; however, the etiology of obesity associated type 2 diabetes mellitus remains largely unknown. There is an urgent need to further broaden the understanding of the molecular mechanism associated in obesity associated type 2 diabetes mellitus. Methods: To screen the differentially expressed genes (DEGs) that might play essential roles in obesity associated type 2 diabetes mellitus, the publicly available expression profiling by high throughput sequencing data (GSE143319) was downloaded and screened for DEGs. Then, Gene Ontology (GO) and REACTOME pathway enrichment analysis were performed. The protein - protein interaction network, miRNA - target genes regulatory network and TF-target gene regulatory network were constructed and analyzed for identification of hub and target genes. The hub genes were validated by receiver operating characteristic (ROC) curve analysis and RT- PCR analysis. Finally, a molecular docking study was performed on over expressed proteins to predict the target small drug molecules. Results: A total of 820 DEGs were identified between -

Multi-Ethnic Genome-Wide Association Study for Atrial Fibrillation Carolina Roselli, Broad Institute of MIT and Harvard Mark D
Multi-ethnic genome-wide association study for atrial fibrillation Carolina Roselli, Broad Institute of MIT and Harvard Mark D. Chaffin, Broad Institute of MIT and Harvard Lu-Chen Weng, Broad Institute of MIT and Harvard Stefanie Aeschbacher, University Hospital Basel Gustav Ahlberg, Copenhagen University Hospital Christine M. Albert, Brigham & Womens Hospital Peter Almgren, Lund University Alvaro Alonso, Emory University Christopher D. Anderson, Broad Institute of MIT and Harvard Krishna G. Aragam, Broad Institute of MIT and Harvard Only first 10 authors above; see publication for full author list. Journal Title: Nature Genetics Volume: Volume 50, Number 9 Publisher: Nature Research (part of Springer Nature) | 2018-09-01, Pages 1225-+ Type of Work: Article | Post-print: After Peer Review Publisher DOI: 10.1038/s41588-018-0133-9 Permanent URL: https://pid.emory.edu/ark:/25593/tdhf0 Final published version: http://dx.doi.org/10.1038/s41588-018-0133-9 Copyright information: © 2018 The Author(s) Accessed October 2, 2021 3:10 AM EDT HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Nat Genet Manuscript Author . Author manuscript; Manuscript Author available in PMC 2018 September 13. Published in final edited form as: Nat Genet. 2018 September ; 50(9): 1225–1233. doi:10.1038/s41588-018-0133-9. Multi-Ethnic Genome-wide Association Study for Atrial Fibrillation A full list of authors and affiliations appears at the end of the article. # These authors contributed equally to this work. Abstract Atrial fibrillation (AF) affects over 33 million individuals worldwide1 and has a complex heritability.2 We conducted the largest meta-analysis of genome-wide association studies for AF to Corresponding author: Patrick T. -

Looking for Missing Proteins in the Proteome Of
Looking for Missing Proteins in the Proteome of Human Spermatozoa: An Update Yves Vandenbrouck, Lydie Lane, Christine Carapito, Paula Duek, Karine Rondel, Christophe Bruley, Charlotte Macron, Anne Gonzalez de Peredo, Yohann Coute, Karima Chaoui, et al. To cite this version: Yves Vandenbrouck, Lydie Lane, Christine Carapito, Paula Duek, Karine Rondel, et al.. Looking for Missing Proteins in the Proteome of Human Spermatozoa: An Update. Journal of Proteome Research, American Chemical Society, 2016, 15 (11), pp.3998-4019. 10.1021/acs.jproteome.6b00400. hal-02191502 HAL Id: hal-02191502 https://hal.archives-ouvertes.fr/hal-02191502 Submitted on 19 Mar 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Journal of Proteome Research 1 2 3 Looking for missing proteins in the proteome of human spermatozoa: an 4 update 5 6 Yves Vandenbrouck1,2,3,#,§, Lydie Lane4,5,#, Christine Carapito6, Paula Duek5, Karine Rondel7, 7 Christophe Bruley1,2,3, Charlotte Macron6, Anne Gonzalez de Peredo8, Yohann Couté1,2,3, 8 Karima Chaoui8, Emmanuelle Com7, Alain Gateau5, AnneMarie Hesse1,2,3, Marlene 9 Marcellin8, Loren Méar7, Emmanuelle MoutonBarbosa8, Thibault Robin9, Odile Burlet- 10 Schiltz8, Sarah Cianferani6, Myriam Ferro1,2,3, Thomas Fréour10,11, Cecilia Lindskog12,Jérôme 11 1,2,3 7,§ 12 Garin , Charles Pineau . -

Gene Ontology Functional Annotations and Pleiotropy
Network based analysis of genetic disease associations Sarah Gilman Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy under the Executive Committee of the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2014 © 2013 Sarah Gilman All Rights Reserved ABSTRACT Network based analysis of genetic disease associations Sarah Gilman Despite extensive efforts and many promising early findings, genome-wide association studies have explained only a small fraction of the genetic factors contributing to common human diseases. There are many theories about where this “missing heritability” might lie, but increasingly the prevailing view is that common variants, the target of GWAS, are not solely responsible for susceptibility to common diseases and a substantial portion of human disease risk will be found among rare variants. Relatively new, such variants have not been subject to purifying selection, and therefore may be particularly pertinent for neuropsychiatric disorders and other diseases with greatly reduced fecundity. Recently, several researchers have made great progress towards uncovering the genetics behind autism and schizophrenia. By sequencing families, they have found hundreds of de novo variants occurring only in affected individuals, both large structural copy number variants and single nucleotide variants. Despite studying large cohorts there has been little recurrence among the genes implicated suggesting that many hundreds of genes may underlie these complex phenotypes. The question -
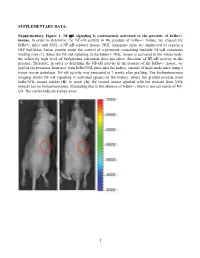
Activation of NF-Κb Signaling Promotes Prostate Cancer Progression in the Mouse and Predicts Poor Progression and Death in Pati
SUPPLEMENTARY DATA: Supplementary Figure 1. NF-B signaling is continuously activated in the prostate of - mouse. In order to determine the NF- '- mouse, we crossed the '- mice with NGL, a NF-B reporter mouse. NGL transgenic mice are engineered to express a GFP/luciferase fusion protein under the control of a promoter containing multiple NF-B consensus binding sites (1). Since the NF-'- NGL mouse is activated in the whole body, the relatively high level of background activation does not allow detection of NF- prostate. Therefore, in order to determine the NF-the '- mouse, we grafted the prostates from 'o the kidney capsule of male nude mice using a tissue rescue technique. NF-B activity was measured at 7 weeks after grafting. The bioluminescence imaging shows NF-B signaling is activated (green) in the kidney, where the grafted prostate from ' use resides (B). In panel (A), the control mouse (grafted with the prostate from NGL mouse) has no bioluminescence, illustrating that in the absence of '-, there is not activation of NF- B. The circles indicate kidney areas. 1 Supplementary Figure 2. NF-B signaling activated in the prostate of Myc/IB bigenic mouse. The prostates from Myc alone (Myc) and bigenic (Myc/IB) mice were harvested at 6 months of age. Activation of NF-B signaling in the prostate was determined by IHC staining of p65-pho antibody. 2 Supplementary Figure 3. Continuous activation of NF-B signaling promotes PCa progression in the Hi-Myc transgenic mouse. The prostates from Myc alone (Myc) and bigeneic (Myc/IB) mice were harvested at 6 months of age. -

Product Name: C1orf185 Polyclonal Antibody, Cy5.5 Conjugated Catalog No
Product Name: C1orf185 Polyclonal Antibody, Cy5.5 Conjugated Catalog No. : TAP01-84955R-Cy5.5 Intended Use: For Research Use Only. Not for used in diagnostic procedures. Size 100ul Concentration 1ug/ul Gene ID 284546 ISO Type Rabbit IgG Clone N/A Immunogen Range Conjugation Cy5.5 Subcellular Locations Applications IF(IHC-P) Cross Reactive Species Human Source KLH conjugated synthetic peptide derived from human C1orf185 Applications with IF(IHC-P)(1:50-200) Dilutions Purification Purified by Protein A. Background Chromosome 1 is the largest human chromosome spanning about 260 million base pairs and making up 8% of the human genome. There are about 3,000 genes on chromosome 1, and considering the great number of genes there are also a large number of diseases associated with chromosome 1. Notably, the rare aging disease Hutchinson-Gilford progeria is associated with the LMNA gene which encodes lamin A. When defective, the LMNA gene product can build up in the nucleus and cause characteristic nuclear blebs. The mechanism of rapidly enhanced aging is unclear and is a topic of continuing exploration. The MUTYH gene is located on chromosome 1 and is partially responsible for familial adenomatous polyposis. Stickler syndrome, Parkinsons, Gaucher disease and Usher syndrome are also associated with chromosome 1. A breakpoint has been identified in 1q which disrupts the DISC1 gene and is linked to schizophrenia. Aberrations in chromosome 1 are found in a variety of cancers including head and neck cancer, malignant melanomaand multiple myeloma. The C1orf185 gene product has been provisionally designated C1orf185 pending further characterization. Synonyms C1orf185; CA185_HUMAN; Chromosome 1 open reading frame 185; Uncharacterized protein C1orf185; Storage Aqueous buffered solution containing 1% BSA, 50% glycerol and 0.09% sodium azide. -

Table S1. 103 Ferroptosis-Related Genes Retrieved from the Genecards
Table S1. 103 ferroptosis-related genes retrieved from the GeneCards. Gene Symbol Description Category GPX4 Glutathione Peroxidase 4 Protein Coding AIFM2 Apoptosis Inducing Factor Mitochondria Associated 2 Protein Coding TP53 Tumor Protein P53 Protein Coding ACSL4 Acyl-CoA Synthetase Long Chain Family Member 4 Protein Coding SLC7A11 Solute Carrier Family 7 Member 11 Protein Coding VDAC2 Voltage Dependent Anion Channel 2 Protein Coding VDAC3 Voltage Dependent Anion Channel 3 Protein Coding ATG5 Autophagy Related 5 Protein Coding ATG7 Autophagy Related 7 Protein Coding NCOA4 Nuclear Receptor Coactivator 4 Protein Coding HMOX1 Heme Oxygenase 1 Protein Coding SLC3A2 Solute Carrier Family 3 Member 2 Protein Coding ALOX15 Arachidonate 15-Lipoxygenase Protein Coding BECN1 Beclin 1 Protein Coding PRKAA1 Protein Kinase AMP-Activated Catalytic Subunit Alpha 1 Protein Coding SAT1 Spermidine/Spermine N1-Acetyltransferase 1 Protein Coding NF2 Neurofibromin 2 Protein Coding YAP1 Yes1 Associated Transcriptional Regulator Protein Coding FTH1 Ferritin Heavy Chain 1 Protein Coding TF Transferrin Protein Coding TFRC Transferrin Receptor Protein Coding FTL Ferritin Light Chain Protein Coding CYBB Cytochrome B-245 Beta Chain Protein Coding GSS Glutathione Synthetase Protein Coding CP Ceruloplasmin Protein Coding PRNP Prion Protein Protein Coding SLC11A2 Solute Carrier Family 11 Member 2 Protein Coding SLC40A1 Solute Carrier Family 40 Member 1 Protein Coding STEAP3 STEAP3 Metalloreductase Protein Coding ACSL1 Acyl-CoA Synthetase Long Chain Family Member 1 Protein -

Rabbit Anti-C1orf185/FITC Conjugated Antibody
SunLong Biotech Co.,LTD Tel: 0086-571- 56623320 Fax:0086-571- 56623318 E-mail:[email protected] www.sunlongbiotech.com Rabbit Anti-C1orf185/FITC Conjugated antibody SL15045R-FITC Product Name: Anti-C1orf185/FITC Chinese Name: FITC标记的1号染色体开放阅读框185抗体 C1orf185; CA185_HUMAN; Chromosome 1 open reading frame 185; Uncharacterized Alias: protein C1orf185; Organism Species: Rabbit Clonality: Polyclonal React Species: Human,Dog,Rabbit, ICC=1:50-200IF=1:50-200 Applications: not yet tested in other applications. optimal dilutions/concentrations should be determined by the end user. Molecular weight: 22kDa Form: Lyophilized or Liquid Concentration: 1mg/ml immunogen: KLH conjugated synthetic peptide derived from human C1orf185 Lsotype: IgG Purification: affinity purified by Protein A Storage Buffer: 0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol. Storewww.sunlongbiotech.com at -20 °C for one year. Avoid repeated freeze/thaw cycles. The lyophilized antibody is stable at room temperature for at least one month and for greater than a year Storage: when kept at -20°C. When reconstituted in sterile pH 7.4 0.01M PBS or diluent of antibody the antibody is stable for at least two weeks at 2-4 °C. background: Chromosome 1 is the largest human chromosome spanning about 260 million base pairs and making up 8% of the human genome. There are about 3,000 genes on chromosome 1, and considering the great number of genes there are also a large Product Detail: number of diseases associated with chromosome 1. Notably, the rare aging disease Hutchinson-Gilford progeria is associated with the LMNA gene which encodes lamin A. -

Current Genomics in Cardiovascular Medicine
446 Current Genomics, 2012, 13, 446-462 Current Genomics in Cardiovascular Medicine 1,2, 3 4 1,2 Vinit Sawhney *, Scott Brouilette , Dominic Abrams , Richard Schilling and Benjamin O’Brien2,5,6,7 1Department of Cardiology, St Bartholomew’s Hospital, London, U.K; 2William Harvey Research Institute, Queen Mary University London, U.K; 3Partek Inc, St Louis, Missouri, USA; 4Department of Cardiology, Children’s Hospital, Boston, USA; 5Department of Intensive Care Medicine and Cardiac Anaesthesia, St Bartholomew’s Hospital, London, U.K; 6 Department of Anaesthesiology, Berlin Heart Institute, Berlin, Germany; 7Life Sciences Division, E.O. Lawrence Ber- keley National Laboratories, Berkeley, California, USA Abstract: Cardiovascular disease (CVD) is a heterogeneous, complex trait that has a major impact on human morbidity and mortality. Common genetic variation may predispose to common forms of CVD in the community, and rare genetic conditions provide unique pathogenetic insights into these diseases. With the advent of the Human Genome Project and the genomic era, new tools and methodologies have revolutionised the field of genetic research in cardiovascular medi- cine. In this review, we describe the rationale for the current emphasis on large-scale genomic studies, elaborate on ge- nome wide association studies and summarise the impact of genomics on clinical cardiovascular medicine and how this may eventually lead to new therapeutics and personalised medicine. Received on: September 15, 2011- Revised on: November 09, 2011- Accepted on: June 13, 2012 Keywords: Cardiovascular disease, GWAS, Gene sequencing, Personalised medicine. INTRODUCTION primary focus on Mendelian disorders to a current emphasis on genome-wide association studies: GWAS. Cardiovascular disease: CVD is a heterogeneous, com- plex trait that has a major impact on human morbidity and However, the clinical practice of cardiovascular medicine mortality. -

Product Name: C1orf185 Polyclonal Antibody, ALEXA FLUOR® 555 Conjugated Catalog No
Product Name: C1orf185 Polyclonal Antibody, ALEXA FLUOR® 555 Conjugated Catalog No. : TAP01-84955R-A555 Intended Use: For Research Use Only. Not for used in diagnostic procedures. Size 100ul Concentration 1ug/ul Gene ID 284546 ISO Type Rabbit IgG Clone N/A Immunogen Range Conjugation ALEXA FLUOR® 555 Subcellular Locations Applications IF(IHC-P) Cross Reactive Species Human Source KLH conjugated synthetic peptide derived from human C1orf185 Applications with IF(IHC-P)(1:50-200) Dilutions Purification Purified by Protein A. Background Chromosome 1 is the largest human chromosome spanning about 260 million base pairs and making up 8% of the human genome. There are about 3,000 genes on chromosome 1, and considering the great number of genes there are also a large number of diseases associated with chromosome 1. Notably, the rare aging disease Hutchinson-Gilford progeria is associated with the LMNA gene which encodes lamin A. When defective, the LMNA gene product can build up in the nucleus and cause characteristic nuclear blebs. The mechanism of rapidly enhanced aging is unclear and is a topic of continuing exploration. The MUTYH gene is located on chromosome 1 and is partially responsible for familial adenomatous polyposis. Stickler syndrome, Parkinsons, Gaucher disease and Usher syndrome are also associated with chromosome 1. A breakpoint has been identified in 1q which disrupts the DISC1 gene and is linked to schizophrenia. Aberrations in chromosome 1 are found in a variety of cancers including head and neck cancer, malignant melanomaand multiple myeloma. The C1orf185 gene product has been provisionally designated C1orf185 pending further characterization. Synonyms C1orf185; CA185_HUMAN; Chromosome 1 open reading frame 185; Uncharacterized protein C1orf185; Storage Aqueous buffered solution containing 1% BSA, 50% glycerol and 0.09% sodium azide.