Population Structure of Denison's Barb, Puntius Denisonii
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

KERALA SOLID WASTE MANAGEMENT PROJECT (KSWMP) with Financial Assistance from the World Bank
KERALA SOLID WASTE MANAGEMENT Public Disclosure Authorized PROJECT (KSWMP) INTRODUCTION AND STRATEGIC ENVIROMENTAL ASSESSMENT OF WASTE Public Disclosure Authorized MANAGEMENT SECTOR IN KERALA VOLUME I JUNE 2020 Public Disclosure Authorized Prepared by SUCHITWA MISSION Public Disclosure Authorized GOVERNMENT OF KERALA Contents 1 This is the STRATEGIC ENVIRONMENTAL ASSESSMENT OF WASTE MANAGEMENT SECTOR IN KERALA AND ENVIRONMENTAL AND SOCIAL MANAGEMENT FRAMEWORK for the KERALA SOLID WASTE MANAGEMENT PROJECT (KSWMP) with financial assistance from the World Bank. This is hereby disclosed for comments/suggestions of the public/stakeholders. Send your comments/suggestions to SUCHITWA MISSION, Swaraj Bhavan, Base Floor (-1), Nanthancodu, Kowdiar, Thiruvananthapuram-695003, Kerala, India or email: [email protected] Contents 2 Table of Contents CHAPTER 1. INTRODUCTION TO THE PROJECT .................................................. 1 1.1 Program Description ................................................................................. 1 1.1.1 Proposed Project Components ..................................................................... 1 1.1.2 Environmental Characteristics of the Project Location............................... 2 1.2 Need for an Environmental Management Framework ........................... 3 1.3 Overview of the Environmental Assessment and Framework ............. 3 1.3.1 Purpose of the SEA and ESMF ...................................................................... 3 1.3.2 The ESMF process ........................................................................................ -

Rain 11 08 2019.Xlsx
Rainfall in 'mm' on 11.08.2019 District River Basin Station Name 11-08-2019 Alappuzha Achencovil Kollakadavu 55.2 Alappuzha Manimala Ambalapuzha 99.3 Alappuzha Muvattupuzha Arookutty 114.4 Alappuzha Muvattupuzha Cherthala 108 Cannanore Anjarakandy Cheruvanchery 96 Cannanore Anjarakandy F.c.s. Pazhassi 93 Cannanore Anjarakandy Kottiyoor 176 Cannanore Anjarakandy Kannavam 72 Cannanore Karaingode Pulingome 167.4 Cannanore Kuppam Alakkode 148.6 Cannanore Peruvamba Kaithaprem 116.2 Cannanore Peruvamba Olayampadi 144.6 Cannanore Ramapuram Cheruthazham 70.2 Cannanore Anjarakandy Maloor 104 Cannanore Valapattanam Mangattuparamba 58.6 Cannanore Anjarakandy Nedumpoil 77.2 Cannanore Valapattanam Palappuzha 80 Cannanore Valapattanam Payyavoor 140 Cannanore Kuppam Alakkode 148.6 Cannanore Valapattanam Thillenkeri 121 Ernakulam Muvattupuzha Piravam 87.2 Ernakulam Periyar Aluva 112.5 Ernakulam Periyar Boothathankettu 79.6 Ernakulam Periyar Keerampara 63.2 Ernakulam Periyar Neriyamangalam 69.8 Idukki Manimala Boyce estate 47 Idukki Muvattupuzha Vannapuram 54.3 Idukki Pambar Marayoor 5.6 Idukki Periyar Chinnar 37 Idukki Periyar FCS Painavu 32.4 Idukki Periyar Kumali 27 Idukki Periyar Nedumkandam 23.8 Idukki Periyar Vandanmedu 34.8 Kasaragod Chandragiri Vidhyanagar 161.8 Kasaragod Chandragiri Kalliyot 142.3 Kasaragod Chandragiri Padiyathadukka 126.4 Kasaragod Karaingode Kakkadavue(cheemeni)fcs 141.8 Kasaragod Manjeswar Manjeswaram 74 Kasaragod Morgal Madhur 145.2 Kasaragod Nileswar Erikkulam 127.4 Kasaragod Shiriya Paika 137 Kasaragod Uppala Uppala 90.5 -

Classic Kerala
Classic Kerala Classic Kerala 9 Days | Kochi to Kochi PRIVATE TOUR: Combining historic • Touring and excursions as per itinerary Cochin takes you through streets still graced Kochi, the hill station of Munnar • Boat ride on Lake Periyar OR visit to a tea with traces of Portuguese, Dutch and British and a houseboat cruise on the plantation colonial architecture. Afternoon/evening free. • Services of English-speaking Indian Overnight - Kochi (B) backwaters of Kerala. In nine specialist guides for all included sight- sunfilled days feast upon the seeing Day 3 : Munnar amazing natural wonders, wildlife, • Entrance fees to all included sights beauty and diversity of stunning • An airport arrival transfer day 1 and a Kerala - God's own country. departure transfer day 9 • All relevant transfer and transportation in private modern Chauffeur driven air- HIGHLIGHTS AND INCLUSIONS conditioned vehicles Trip Highlights What's Not Included • Historic Kochi - Parsi Synagogue, Dutch • International flights and visas Palace, St Francis Church, Fort Cochin, • Tipping - An entirely personal gesture Kochi - Munnar. A drive east to to Munnar colourful boats and Chinese fishing nets - a quaint hill station nestled on the verdant on the waterfront ITINERARY slopes of tea plantations, that exudes a rare • Munnar Hill Station - The summer retreat old world charm and reminds one of the days of the British during colonial times Day 1 : Kochi of the Raj in its elegant splendour. At a height • The protected Periyar Wildlife Sanctuary Once you arrive Kochi you'll be met at the of 1525 metres, Munnar offers breathtaking and lake - Elephant and tiger reserve airport and transferred to your hotel. -

Smujo International
BIODIVERSITAS ISSN: 1412-033X Volume 21, Number 3, March 2020 E-ISSN: 2085-4722 Pages: 1196-1200 DOI: 10.13057/biodiv/d210346 Short Communication: Proximate analysis, amino acid profile and albumin concentration of various weights of Giant Snakehead (Channa micropeltes) from Kapuas Hulu, West Kalimantan, Indonesia WAHYU WIRA PRATAMA1, HAPPY NURSYAM2, ANIK MARTINAH HARIATI3, R. ADHARYAN ISLAMY3,, VERYL HASAN4, 1Program of Aquaculture, Faculty of Fisheries and Marine Science, Universitas Brawijaya. Jl. Veteran No.16, Malang 65145, East Java, Indonesia 2Department of Fishery Products Technology, Faculty of Fisheries and Marine Science, Universitas Brawijaya. Jl. Veteran No.16, Malang 65145, East Java, Indonesia 3Departement of Aquaculture, Faculty of Fisheries and Marine Science, Universitas Brawijaya. Jl. Veteran No.16, Malang 65145, East Java, Indonesia. Tel.: +62-341-553-512, Fax.: +62-341-556-837, email: [email protected] 4Department of Fish Health Management and Aquaculture, Faculty of Fisheries and Marine Science, Universitas Airlangga. Kampus C Unair, Jl. Mulyosari, Surabaya 60113, East Java, Indonesia. Tel.: +62-31-315911541, Fax.: +62-31-5965741, email: [email protected] Manuscript received: 13 November 2019. Revision accepted: 23 February 2020. Abstract. Pratama WW, Nursyam H, Hariati AM, Islamy RA, Hasan V. 2020. Short Communication: Proximate analysis, amino acid profile and albumin concentration of various weights of Giant Snakehead (Channa micropeltes) from Kapuas Hulu, West Kalimantan, Indonesia. Biodiversitas 21: 1196-1200. Fish is an important foodstuff due to its nutritional value and high protein. One of popular fish as a foodstuff in tropical Asia is giant snakehead fish (Channa micropeltes). This study aims to examine the proximate composition, amino acid profile, and albumin concentration of giant snakeheads in various weights and to determine the best weight of giant snakeheads according to the proximate, amino acid, and albumin concentration. -

Snakeheadsnepal Pakistan − (Pisces,India Channidae) PACIFIC OCEAN a Biologicalmyanmar Synopsis Vietnam
Mongolia North Korea Afghan- China South Japan istan Korea Iran SnakeheadsNepal Pakistan − (Pisces,India Channidae) PACIFIC OCEAN A BiologicalMyanmar Synopsis Vietnam and Risk Assessment Philippines Thailand Malaysia INDIAN OCEAN Indonesia Indonesia U.S. Department of the Interior U.S. Geological Survey Circular 1251 SNAKEHEADS (Pisces, Channidae)— A Biological Synopsis and Risk Assessment By Walter R. Courtenay, Jr., and James D. Williams U.S. Geological Survey Circular 1251 U.S. DEPARTMENT OF THE INTERIOR GALE A. NORTON, Secretary U.S. GEOLOGICAL SURVEY CHARLES G. GROAT, Director Use of trade, product, or firm names in this publication is for descriptive purposes only and does not imply endorsement by the U.S. Geological Survey. Copyrighted material reprinted with permission. 2004 For additional information write to: Walter R. Courtenay, Jr. Florida Integrated Science Center U.S. Geological Survey 7920 N.W. 71st Street Gainesville, Florida 32653 For additional copies please contact: U.S. Geological Survey Branch of Information Services Box 25286 Denver, Colorado 80225-0286 Telephone: 1-888-ASK-USGS World Wide Web: http://www.usgs.gov Library of Congress Cataloging-in-Publication Data Walter R. Courtenay, Jr., and James D. Williams Snakeheads (Pisces, Channidae)—A Biological Synopsis and Risk Assessment / by Walter R. Courtenay, Jr., and James D. Williams p. cm. — (U.S. Geological Survey circular ; 1251) Includes bibliographical references. ISBN.0-607-93720 (alk. paper) 1. Snakeheads — Pisces, Channidae— Invasive Species 2. Biological Synopsis and Risk Assessment. Title. II. Series. QL653.N8D64 2004 597.8’09768’89—dc22 CONTENTS Abstract . 1 Introduction . 2 Literature Review and Background Information . 4 Taxonomy and Synonymy . -

Length-Weight and Length-Length Relationship of Three Species of Snakehead Fish, Channa Diplogramma, C
Journal of Threatened Taxa | www.threatenedtaxa.org | 26 September 2013 | 5(13): 4769–4773 Western Ghats Special Series ISSN Length-weight and length-length relationship of three Communication Short Online 0974–7907 species of snakehead fish, Channa diplogramma, C. marulius Print 0974–7893 and C. striata from the riverine reaches of Lake Vembanad, OPEN ACCESS Kerala, India Anvar Ali 1, Neelesh Dahanukar 2 & Rajeev Raghavan 3 1,3 Conservation Research Group (CRG), St. Albert’s College, Kochi, Kerala 682018, India 2 Indian Institute of Science Education and Research (IISER), Dr. Homi Bhabha Road, Pashan, Pune, Maharashtra 411008, India 2,3 Zoo Outreach Organization (ZOO), 96 Kumudham Nagar, Vilankurichi Road, Coimbatore, Tamil Nadu 641035, India 1 [email protected], 2 [email protected], 3 [email protected] (corresponding author) Abstract: The length-weight relationship (LWR) and length-length Snakeheads of the genus Channa Scopoli, 1777, are relationships (LLR) of three snakehead fishes, Channa diplogramma, among the most popular food fishes in tropical Asia (Wee C. marulius and C. striata, exploited by small-scale fishers in the riverine reaches of Lake Vembanad, Kerala were studied using the 1982). In addition to being a common staple food fish, allometric growth equation Y = aXb. Our analysis shows that the LWR snakeheads are also consumed therapeutically for wound of C. diplogramma and C. marulius is nonisometric with exponents much smaller than the cubic value (b = 3), while that of C. striata is healing and reducing post-operative pain and discomfort isometric. Channa marulius showed a definite change in LWR with (Gam et al. -

International Journal of Engineering Sciences &Research Technology
IJESRT: 9(6), June, 2020 ISSN: 2277-9655 International Journal of Engineering Sciences &Research Technology (A Peer Reviewed Online Journal) Impact Factor: 5.164 IJESRT Chief Editor Executive Editor Dr. J.B. Helonde Mr. SomilMayurShah Website:www.ijesrt.com Mail:[email protected] ISSN: 2277-9655 [Georgeet al., 9(6): June, 2020] Impact Factor: 5.164 IC™ Value: 3.00 CODEN: IJESS7 IJESRT INTERNATIONAL JOURNAL OF ENGINEERING SCIENCES & RESEARCH TECHNOLOGY HYDROLOGIC BEHAVIOUR OF HIGHLAND CATCHMENT IN HUMID TROPICAL REGION USING QSWAT MODEL Celine George*1& Haritha P. Scaria 2 *1Principal Scientist & Head, Centre for Water Resources Development and Management (CWRDM) Sub Centre, Manimalakunnu, Koothattukulam, Kerala – 686 662, India 2B.Tech (Civil), National Institute of Technology, Kozhikode, Kerala, India DOI: https://doi.org/10.29121/ijesrt.v9.i6.2020.16 ABSTRACT Water resources planning and management of a region requires an understanding of the water balance in the region. The Soil and Water Assessment Tool (SWAT) with QGIS interface (QSWAT) has been used here to arrive at the water balance components in the Palapuzha watershed of Valapattanam river basin in Kerala. Valapattanam river drains an area of 1867 sq.km. with 456 sq.km. area in Karnataka State. The river basin receives an average annual rainfall of 3600 mm. The Palapuzha watershed drains an area of 237.25 sq.km with an average annual rainfall of 4562 mm. The QSWAT model has been calibrated and validated using data for a period of eight years (2000-2007) for which both rainfall and streamflow data are available. The model was successful in simulating monthly streamflow during the calibration and validation periods with Nash Sutcliffe efficiency and correlation co-efficient greater than 0.75 and percent bias less than 10%, showing that the model is very good for predicting streamflow in Valapattanam river basin. -

A CONCISE REPORT on BIODIVERSITY LOSS DUE to 2018 FLOOD in KERALA (Impact Assessment Conducted by Kerala State Biodiversity Board)
1 A CONCISE REPORT ON BIODIVERSITY LOSS DUE TO 2018 FLOOD IN KERALA (Impact assessment conducted by Kerala State Biodiversity Board) Editors Dr. S.C. Joshi IFS (Rtd.), Dr. V. Balakrishnan, Dr. N. Preetha Editorial Board Dr. K. Satheeshkumar Sri. K.V. Govindan Dr. K.T. Chandramohanan Dr. T.S. Swapna Sri. A.K. Dharni IFS © Kerala State Biodiversity Board 2020 All rights reserved. No part of this book may be reproduced, stored in a retrieval system, tramsmitted in any form or by any means graphics, electronic, mechanical or otherwise, without the prior writted permission of the publisher. Published By Member Secretary Kerala State Biodiversity Board ISBN: 978-81-934231-3-4 Design and Layout Dr. Baijulal B A CONCISE REPORT ON BIODIVERSITY LOSS DUE TO 2018 FLOOD IN KERALA (Impact assessment conducted by Kerala State Biodiversity Board) EdItorS Dr. S.C. Joshi IFS (Rtd.) Dr. V. Balakrishnan Dr. N. Preetha Kerala State Biodiversity Board No.30 (3)/Press/CMO/2020. 06th January, 2020. MESSAGE The Kerala State Biodiversity Board in association with the Biodiversity Management Committees - which exist in all Panchayats, Municipalities and Corporations in the State - had conducted a rapid Impact Assessment of floods and landslides on the State’s biodiversity, following the natural disaster of 2018. This assessment has laid the foundation for a recovery and ecosystem based rejuvenation process at the local level. Subsequently, as a follow up, Universities and R&D institutions have conducted 28 studies on areas requiring attention, with an emphasis on riverine rejuvenation. I am happy to note that a compilation of the key outcomes are being published. -

Fcover-RR114-High
RESEARCH REPORT Developing Procedures for 114 Assessment of Ecological Status of Indian River Basins in the Context of Environmental Water Requirements Vladimir Smakhtin, Muthukumarasamy Arunachalam, Sandeep Behera, Archana Chatterjee, Srabani Das, Parikshit Gautam, Gaurav D. Joshi, Kumbakonam G. Sivaramakrishnan and K. Sankaran Unni International Water Management IWMI is a Future Harvest Center Institute supported by the CGIAR Research Reports IWMI’s mission is to improve water and land resources management for food, livelihoods and nature. In serving this mission, IWMI concentrates on the integration of policies, technologies and management systems to achieve workable solutions to real problems—practical, relevant results in the field of irrigation and water and land resources. The publications in this series cover a wide range of subjects—from computer modeling to experience with water user associations—and vary in content from directly applicable research to more basic studies, on which applied work ultimately depends. Some research reports are narrowly focused, analytical and detailed empirical studies; others are wide-ranging and synthetic overviews of generic problems. Although most of the reports are published by IWMI staff and their collaborators, we welcome contributions from others. Each report is reviewed internally by IWMI’s own staff and Fellows, and by external reviewers. The reports are published and distributed both in hard copy and electronically (www.iwmi.org) and where possible all data and analyses will be available as separate downloadable files. Reports may be copied freely and cited with due acknowledgment. Research Report 114 Developing Procedures for Assessment of Ecological Status of Indian River Basins in the Context of Environmental Water Requirements Vladimir Smakhtin, Muthukumarasamy Arunachalam, Sandeep Behera, Archana Chatterjee, Srabani Das, Parikshit Gautam, Gaurav D. -

Periyar and Pamba River Basin
Periyar and Pamba River Basin The Periyar River with a length of 228 km is the second longest river of this basin. It rises from the forest –clad Sivagiri peak, 80 km. south of Devikulam at an elevation of 2,438 m above sea level. The total drainage area of the river is 5,243 sq. km., out of which 113 sq. km. lies in Tamil Nadu. The Pamba is the third longest river of the basin. It is 177 Km. in length with the total drainage area spread of 1961 sq. km. It is formed by the confluence of the rivers Pamba Aar, Kakki Aar, Arudai Aar, Kakkad Aar and kali Aar. Area: Periyar and others Sub Basin consists an area of 21895.21 sq. km. Watershed: Periyar and others sub basin contains of 37 watersheds with size range of 341 – 934 sq. km. Population: 14162844 in 14 districts. Agro ecological zone: Hot humid perhumid ecoregion with red, lateritic and alluvium- derived soils. The other two zones are hot semi-arid eco-region with red loamy soils and the hot sub-humid to semi arid eco-region with coastal alluvium derived soils. Major irrigation projects: Pamba Irrigation Project: This project, located in Pattanamthitta district, aims at the utilization of the waters of Sabarigiri Hydro Electric project for irrigation purpose. The water is let into the river Kakkad, a tributary of Pamba River, and is picked up at Maniyar by a barrage. The water, thus collected is diverted through a canal on the left bank of the river. The project consists of a barrage of length 115.22 m with FRL at 34.62 m. -
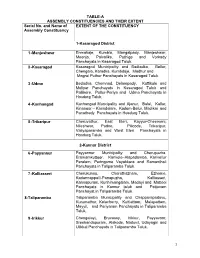
List of Lacs with Local Body Segments (PDF
TABLE-A ASSEMBLY CONSTITUENCIES AND THEIR EXTENT Serial No. and Name of EXTENT OF THE CONSTITUENCY Assembly Constituency 1-Kasaragod District 1 -Manjeshwar Enmakaje, Kumbla, Mangalpady, Manjeshwar, Meenja, Paivalike, Puthige and Vorkady Panchayats in Kasaragod Taluk. 2 -Kasaragod Kasaragod Municipality and Badiadka, Bellur, Chengala, Karadka, Kumbdaje, Madhur and Mogral Puthur Panchayats in Kasaragod Taluk. 3 -Udma Bedadka, Chemnad, Delampady, Kuttikole and Muliyar Panchayats in Kasaragod Taluk and Pallikere, Pullur-Periya and Udma Panchayats in Hosdurg Taluk. 4 -Kanhangad Kanhangad Muncipality and Ajanur, Balal, Kallar, Kinanoor – Karindalam, Kodom-Belur, Madikai and Panathady Panchayats in Hosdurg Taluk. 5 -Trikaripur Cheruvathur, East Eleri, Kayyur-Cheemeni, Nileshwar, Padne, Pilicode, Trikaripur, Valiyaparamba and West Eleri Panchayats in Hosdurg Taluk. 2-Kannur District 6 -Payyannur Payyannur Municipality and Cherupuzha, Eramamkuttoor, Kankole–Alapadamba, Karivellur Peralam, Peringome Vayakkara and Ramanthali Panchayats in Taliparamba Taluk. 7 -Kalliasseri Cherukunnu, Cheruthazham, Ezhome, Kadannappalli-Panapuzha, Kalliasseri, Kannapuram, Kunhimangalam, Madayi and Mattool Panchayats in Kannur taluk and Pattuvam Panchayat in Taliparamba Taluk. 8-Taliparamba Taliparamba Municipality and Chapparapadavu, Kurumathur, Kolacherry, Kuttiattoor, Malapattam, Mayyil, and Pariyaram Panchayats in Taliparamba Taluk. 9 -Irikkur Chengalayi, Eruvassy, Irikkur, Payyavoor, Sreekandapuram, Alakode, Naduvil, Udayagiri and Ulikkal Panchayats in Taliparamba -

Beyond the Tsunami
Beyond the Tsunami Current Status of Mangroves in Kerala and Tamil Nadu, India, with regard to Vegetation, Community Perceptions and Policy 2008 Nibedita Mukherjee, Farid Dahdouh-Guebas The Coastal and Marine Programme at ATREE and Kartik Shanker is interdisciplinary in its approach and applies skills in the natural and social sciences to its United Nations Team for Ashoka Trust for Research in Tsunami Recovery Support Ecology and the Environment research and conservation interventions. Beyond the Tsunami Current Status of Mangroves in Kerala and Tamil Nadu, India with regard to Vegetation, Community Perceptions and Policy Nibedita Mukherjee, Farid Dahdouh-Guebas and Kartik Shanker United Nations Team for Tsunami Recovery Support Author details: Kartik Shanker Adjunct Fellow, Ashoka Trust for Research in Ecology and the Environment (ATREE) 659, 5th Main Road, Hebbal, Bangalore 560092. India. and Assistant Professor, Centre for Ecological Sciences, Indian Institute of Science, Bangalore 560012. India. E-mail: [email protected] Nibedita Mukherjee Research Associate, Ashoka Trust for Research in Ecology and the Environment (ATREE) 659, 5th Main Road, Hebbal, Bangalore 560092. India. and PhD Student, Plant Biology and Nature Management (APNA), Vrije Universiteit Brussel – VUB, Pleinlaan, B-1050 Brussels, Belgium. E-mail: [email protected] Farid Dahdouh-Guebas Complexity and Dynamics of Tropical Systems, Université Libre deBruxelles – ULB, Av. F.D. Roosevelt 50, B-1050 Brussels, Belgium. E-mail: [email protected] Acknowledgements We would like to thank the United Nations Development Programme for funding this initiative. We would also like to thank the Forest Department of Kerala, Andhra Pradesh and Tamil Nadu for helping in the completion of this project.