TCR Signaling Promotes the Assembly of Ranbp2/Rangap1-SUMO1/Ubc9 Nuclear Pore Subcomplex
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

XPO1E571K Mutation Modifies Exportin 1 Localisation And
cancers Article XPO1E571K Mutation Modifies Exportin 1 Localisation and Interactome in B-Cell Lymphoma Hadjer Miloudi 1, Élodie Bohers 1,2, François Guillonneau 3 , Antoine Taly 4,5 , Vincent Cabaud Gibouin 6,7 , Pierre-Julien Viailly 1,2 , Gaëtan Jego 6,7 , Luca Grumolato 8 , Fabrice Jardin 1,2 and Brigitte Sola 1,* 1 INSERM U1245, Unicaen, Normandie University, F-14000 Caen, France; [email protected] (H.M.); [email protected] (E.B.); [email protected] (P.-J.V.); [email protected] (F.J.) 2 Centre de lutte contre le Cancer Henri Becquerel, F-76000 Rouen, France 3 Plateforme Protéomique 3P5, Université de Paris, Institut Cochin, INSERM, CNRS, F-75014 Paris, France; [email protected] 4 Laboratoire de Biochimie Théorique, CNRS UPR 9030, Université de Paris, F-75005 Paris, France; [email protected] 5 Institut de Biologie Physico-Chimique, Fondation Edmond de Rothschild, PSL Research University, F-75005 Paris, France 6 INSERM, LNC UMR1231, F-21000 Dijon, France; [email protected] (V.C.G.); [email protected] (G.J.) 7 Team HSP-Pathies, University of Burgundy and Franche-Comtée, F-21000 Dijon, France 8 INSERM U1239, Unirouen, Normandie University, F-76130 Mont-Saint-Aignan, France; [email protected] * Correspondence: [email protected]; Tel.: +33-2-3156-8210 Received: 11 September 2020; Accepted: 28 September 2020; Published: 30 September 2020 Simple Summary: Almost 25% of patients with either primary mediastinal B-cell lymphoma (PMBL) or classical Hodgkin lymphoma (cHL) possess a recurrent mutation of the XPO1 gene encoding the major nuclear export protein. -

Plant Rangaps Are Localized at the Nuclear Envelope in Interphase and Associated with Microtubules in Mitotic Cells
The Plant Journal (2002) 30(6), 699±709 Plant RanGAPs are localized at the nuclear envelope in interphase and associated with microtubules in mitotic cells Aniko Pay1, Katja Resch2, Hanns Frohnmeyer2, Erzsebet Fejes1, Ferenc Nagy1 and Peter Nick2,* 1Plant Biology Institute, Biological Research Center, H-6701 Szeged, PO Box 521, Hungary, and 2Institut fuÈr Biologie II/Botanik, SchaÈnzlestrasse 1, UniversitaÈt Freiburg, D-79104 Freiburg, Germany Received 17 December 2001; revised 7 March 2002; accepted 15 March 2002. *For correspondence (fax +49 761 203 2612; e-mail [email protected]). Summary In animals and yeast, the small GTP-binding protein Ran has multiple functions ± it is involved in mediating (i) the directional passage of proteins and RNA through the nuclear pores in interphase cells; and (ii) the formation of spindle asters, the polymerization of microtubules, and the re-assembly of the nuclear envelope in mitotic cells. Nucleotide binding of Ran is modulated by a series of accessory proteins. For instance, the hydrolysis of RanGTP requires stimulation by the RanGTPase protein RanGAP. Here we report the complementation of the yeast RanGAP mutant rna1 with Medicago sativa and Arabidopsis thaliana cDNAs encoding RanGAP-like proteins. Confocal laser microscopy of Arabidopsis plants overexpressing chimeric constructs of GFP with AtRanGAP1 and 2 demonstrated that the fusion protein is localized to patchy areas at the nuclear envelope of interphase cells. In contrast, the cellular distribution of RanGAPs in synchronized tobacco cells undergoing mitosis is characteristically different. Double-immuno¯uorescence shows that RanGAPs are co-localized with spindle microtubules during anaphase, with the microtubular phragmoplast and the surface of the daughter nuclei during telophase. -

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

SUMO-1 Regulates the Conformational Dynamics of Thymine-DNA
Smet-Nocca et al. BMC Biochemistry 2011, 12:4 http://www.biomedcentral.com/1471-2091/12/4 RESEARCHARTICLE Open Access SUMO-1 regulates the conformational dynamics of Thymine-DNA Glycosylase regulatory domain and competes with its DNA binding activity Caroline Smet-Nocca1, Jean-Michel Wieruszeski2, Hélène Léger1,3, Sebastian Eilebrecht1,3, Arndt Benecke1,3* Abstract Background: The human thymine-DNA glycosylase (TDG) plays a dual role in base excision repair of G:U/T mismatches and in transcription. Regulation of TDG activity by SUMO-1 conjugation was shown to act on both functions. Furthermore, TDG can interact with SUMO-1 in a non-covalent manner. Results: Using NMR spectroscopy we have determined distinct conformational changes in TDG upon either covalent sumoylation on lysine 330 or intermolecular SUMO-1 binding through a unique SUMO-binding motif (SBM) localized in the C-terminal region of TDG. The non-covalent SUMO-1 binding induces a conformational change of the TDG amino-terminal regulatory domain (RD). Such conformational dynamics do not exist with covalent SUMO-1 attachment and could potentially play a broader role in the regulation of TDG functions for instance during transcription. Both covalent and non-covalent processes activate TDG G:U repair similarly. Surprisingly, despite a dissociation of the SBM/SUMO-1 complex in presence of a DNA substrate, SUMO-1 preserves its ability to stimulate TDG activity indicating that the non-covalent interactions are not directly involved in the regulation of TDG activity. SUMO-1 instead acts, as demonstrated here, indirectly by competing with the regulatory domain of TDG for DNA binding. -

The CARMA3-Bcl10-MALT1 Signalosome Drives NF-Κb Activation and Promotes Aggressiveness in Angiotensin II Receptor-Positive Breast Cancer
Author Manuscript Published OnlineFirst on December 19, 2017; DOI: 10.1158/0008-5472.CAN-17-1089 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Molecular and Cellular Pathobiology .. The CARMA3-Bcl10-MALT1 Signalosome Drives NF-κB Activation and Promotes Aggressiveness in Angiotensin II Receptor-positive Breast Cancer. Prasanna Ekambaram1, Jia-Ying (Lloyd) Lee1, Nathaniel E. Hubel1, Dong Hu1, Saigopalakrishna Yerneni2, Phil G. Campbell2,3, Netanya Pollock1, Linda R. Klei1, Vincent J. Concel1, Phillip C. Delekta4, Arul M. Chinnaiyan4, Scott A. Tomlins4, Daniel R. Rhodes4, Nolan Priedigkeit5,6, Adrian V. Lee5,6, Steffi Oesterreich5,6, Linda M. McAllister-Lucas1,*, and Peter C. Lucas1,* 1Departments of Pathology and Pediatrics, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania 2Department of Biomedical Engineering, Carnegie Mellon University, Pittsburgh, Pennsylvania 3McGowan Institute for Regenerative Medicine, University of Pittsburgh, Pittsburgh, Pennsylvania 4Department of Pathology, University of Michigan Medical School, Ann Arbor, Michigan 5Women’s Cancer Research Center, Magee-Womens Research Institute, University of Pittsburgh Cancer Institute, Pittsburgh, Pennsylvania 6Department of Pharmacology and Chemical Biology, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania Current address for P.C. Delekta: Department of Microbiology & Molecular Genetics, Michigan State University, East Lansing, Michigan Current address for D.R. Rhodes: Strata -

The Drug Sensitivity and Resistance Testing (DSRT) Approach
A phenotypic screening and machine learning platform eciently identifies triple negative breast cancer-selective and readily druggable targets Prson Gautam 1 Alok Jaiswal 1 Tero Aittokallio 1, 2 Hassan Al Ali 3 Krister Wennerberg 1,4 Identifying eective oncogenic targets is challenged by the complexity of genetic alterations in 1Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Finland cancer and their poorly understood relation to cell function and survival. There is a need for meth- Current kinome coverage of kinase inhibitors in TNBC exhibit diverse kinase dependencies MFM-223 is selectively addicted to FGFR2 2Department of Mathematics and Statistics, University of Turku, Finland 3The Miami Project to Cure Paralysis, Peggy and Harold Katz Family Drug Discovery Center, A A Sylvester Comprehensive Cancer Center, and Department of Neurological Surgery and Medicine ods that rapidly and accurately identify “pharmacologically eective” targets without the require- clinical evaluation TN Kinases MFM-223 CAL-120 MDA-MB-231 TNBC TNBC TNBC TNBC TNBC TNBC HER2+ 100 University of Miami Miller School of Medicine, Miami, FL 33136, USA. non- HER2+ FGFR1 0.97 0.00 0.00 MFM-223 BL1 BL2 M MSL IM LAR ER+, PR+ 50 ment for priori knowledge of complex signaling networks. We developed an approach that uses ma- cancerous FGFR2 56.46 0.00 0.00 CAL-120 25 4 MDA-MB-231 Biotech Research & Innovation Centre (BRIC) and Novo Nordisk Foundation Center HCC1937 CAL-85-1 CAL-120 MDA-MB-231 DU4475 CAL-148 MCF-10A SK-BR-3 BT-474 FGFR3 25.10 0.00 0.00 0 chine learning to relate results from unbiased phenotypic screening of kinase inhibitors to their bio- for Stem Cell Biology (DanStem), University of Copenhagen, Denmark HCC1599 HDQ-P1 BT-549 MDA-MB-436 MFM-223 FGFR4 0.00 0.00 0.00 MAXIS*Bk Clinical status MDA-MB-468 CAL-51 Hs578T MDA-MB-453 score chemical activity data. -

The LUBAC Participates in Lysophosphatidic Acid-Induced NF-Κb Activation
bioRxiv preprint doi: https://doi.org/10.1101/2020.02.13.948125; this version posted February 13, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The LUBAC participates in Lysophosphatidic Acid-induced NF-κB Activation Tiphaine Douanne1, Sarah Chapelier1, Robert Rottapel2, Julie Gavard1,3, Nicolas Bidère1,* 1Université de Nantes, INSERM, CNRS, CRCINA, Team SOAP, F-440000 Nantes, France; 2Princess Margaret Cancer Centre, University Health Network, Toronto, Ontario, Canada ; 3Institut de Cancérologie de l’Ouest, Site René Gauducheau, 44800 Saint-Herblain, France *Author for correspondence: [email protected] Abstract The natural bioactive glycerophospholipid lysophosphatidic acid (LPA) binds to its cognate G protein-coupled receptors (GPCRs) on the cell surface to promote the activation of several transcription factors, including NF-κB. LPA-mediated activation of NF-κB relies on the formation of a signalosome that contains the scaffold CARMA3, the adaptor BCL10 and the paracaspase MALT1 (CBM complex). The CBM has been extensively studied in lymphocytes, where it links antigen receptors to NF-κB activation via the recruitment of the linear ubiquitin assembly complex (LUBAC), a tripartite complex of HOIP, HOIL1 and SHARPIN. Moreover, MALT1 cleaves the LUBAC subunit HOIL1 to further enhance NF- κB activation. However, the contribution of the LUBAC downstream of GPCRs has not been investigated. By using murine embryonic fibroblasts from mice deficient for HOIP, HOIL1 and SHARPIN, we report that the LUBAC is crucial for the activation of NF-κB in response to LPA. Further echoing the situation in lymphocytes, LPA unbridles the protease activity of MALT1, which cleaves HOIL1 at the Arginine 165. -

Distinct Basket Nucleoporins Roles in Nuclear Pore Function and Gene Expression
bioRxiv preprint doi: https://doi.org/10.1101/685263; this version posted June 28, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. This article is a US Government work. It is not subject to copyright under 17 USC 105 and is also made available for use under a CC0 license. Distinct Basket Nucleoporins roles in Nuclear Pore Function and Gene Expression: Tpr is an integral component of the TREX-2 mRNA export pathway Vasilisa Aksenova1, Hang Noh Lee1, †, Alexandra Smith1, †, Shane Chen1, †, Prasanna Bhat3, †, James Iben2, Carlos Echeverria1, Beatriz Fontoura3, Alexei Arnaoutov1 and Mary Dasso1, * 1Division of Molecular and Cellular Biology, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892, USA. 2Molecular Genomics Core, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, Maryland 20879 3Department of Cell Biology, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA. † These authors contributed equally to this work. *Correspondence: [email protected]. Acronyms: NPC – nuclear pore complex; BSK-NUPs – basket nucleoporins; NG – NeonGreen; AID - Auxin Inducible Degron 1 bioRxiv preprint doi: https://doi.org/10.1101/685263; this version posted June 28, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. This article is a US Government work. It is not subject to copyright under 17 USC 105 and is also made available for use under a CC0 license. Abstract Nuclear pore complexes (NPCs) are important for many processes beyond nucleocytoplasmic trafficking, including protein modification, chromatin remodeling, transcription, mRNA processing and mRNA export. -

Download the Abstract Book
1 Exploring the male-induced female reproduction of Schistosoma mansoni in a novel medium Jipeng Wang1, Rui Chen1, James Collins1 1) UT Southwestern Medical Center. Schistosomiasis is a neglected tropical disease caused by schistosome parasites that infect over 200 million people. The prodigious egg output of these parasites is the sole driver of pathology due to infection. Female schistosomes rely on continuous pairing with male worms to fuel the maturation of their reproductive organs, yet our understanding of their sexual reproduction is limited because egg production is not sustained for more than a few days in vitro. Here, we explore the process of male-stimulated female maturation in our newly developed ABC169 medium and demonstrate that physical contact with a male worm, and not insemination, is sufficient to induce female development and the production of viable parthenogenetic haploid embryos. By performing an RNAi screen for genes whose expression was enriched in the female reproductive organs, we identify a single nuclear hormone receptor that is required for differentiation and maturation of germ line stem cells in female gonad. Furthermore, we screen genes in non-reproductive tissues that maybe involved in mediating cell signaling during the male-female interplay and identify a transcription factor gli1 whose knockdown prevents male worms from inducing the female sexual maturation while having no effect on male:female pairing. Using RNA-seq, we characterize the gene expression changes of male worms after gli1 knockdown as well as the female transcriptomic changes after pairing with gli1-knockdown males. We are currently exploring the downstream genes of this transcription factor that may mediate the male stimulus associated with pairing. -

The Nuclear Export Factor CRM1 Controls Juxta-Nuclear Microtubule-Dependent Virus Transport I-Hsuan Wang1,*,‡, Christoph J
© 2017. Published by The Company of Biologists Ltd | Journal of Cell Science (2017) 130, 2185-2195 doi:10.1242/jcs.203794 RESEARCH ARTICLE The nuclear export factor CRM1 controls juxta-nuclear microtubule-dependent virus transport I-Hsuan Wang1,*,‡, Christoph J. Burckhardt1,2,‡, Artur Yakimovich1,‡, Matthias K. Morf1,3 and Urs F. Greber1,§ ABSTRACT directly bind to virions, or move virions in endocytic or secretory Transport of large cargo through the cytoplasm requires motor vesicles (Greber and Way, 2006; Marsh and Helenius, 2006; proteins and polarized filaments. Viruses that replicate in the nucleus Yamauchi and Greber, 2016). While short-range transport of virions of post-mitotic cells use microtubules and the dynein–dynactin motor often depends on actin and myosin motors (Taylor et al., 2011), to traffic to the nuclear membrane and deliver their genome through long-range cytoplasmic transport requires microtubules, and, in the nuclear pore complexes (NPCs) into the nucleus. How virus particles case of incoming virions, the minus-end-directed motor complex – (virions) or cellular cargo are transferred from microtubules to the dynein dynactin (Dodding and Way, 2011; Hsieh et al., 2010). NPC is unknown. Here, we analyzed trafficking of incoming Like cellular cargo, viruses engage in bidirectional motor- cytoplasmic adenoviruses by single-particle tracking and super- dependent motility by recruiting cytoskeletal motors of opposite resolution microscopy. We provide evidence for a regulatory role of directionality (Greber and Way, 2006; Scherer and Vallee, 2011; CRM1 (chromosome-region-maintenance-1; also known as XPO1, Welte, 2004). The net transport balance of a cargo is determined at exportin-1) in juxta-nuclear microtubule-dependent adenovirus the level of motor recruitment, motor coordination and the tug-of- transport. -
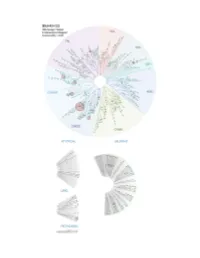
Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) BSJ-03-123 AAK1 AAK1 94 1000 BSJ-03-123 ABL1(E255K)-phosphorylated ABL1 79 1000 BSJ-03-123 ABL1(F317I)-nonphosphorylated ABL1 89 1000 BSJ-03-123 ABL1(F317I)-phosphorylated ABL1 98 1000 BSJ-03-123 ABL1(F317L)-nonphosphorylated ABL1 86 1000 BSJ-03-123 ABL1(F317L)-phosphorylated ABL1 89 1000 BSJ-03-123 ABL1(H396P)-nonphosphorylated ABL1 76 1000 BSJ-03-123 ABL1(H396P)-phosphorylated ABL1 90 1000 BSJ-03-123 ABL1(M351T)-phosphorylated ABL1 100 1000 BSJ-03-123 ABL1(Q252H)-nonphosphorylated ABL1 56 1000 BSJ-03-123 ABL1(Q252H)-phosphorylated ABL1 97 1000 BSJ-03-123 ABL1(T315I)-nonphosphorylated ABL1 100 1000 BSJ-03-123 ABL1(T315I)-phosphorylated ABL1 85 1000 BSJ-03-123 ABL1(Y253F)-phosphorylated ABL1 100 1000 BSJ-03-123 ABL1-nonphosphorylated ABL1 60 1000 BSJ-03-123 ABL1-phosphorylated ABL1 79 1000 BSJ-03-123 ABL2 ABL2 89 1000 BSJ-03-123 ACVR1 ACVR1 100 1000 BSJ-03-123 ACVR1B ACVR1B 95 1000 BSJ-03-123 ACVR2A ACVR2A 100 1000 BSJ-03-123 ACVR2B ACVR2B 96 1000 BSJ-03-123 ACVRL1 ACVRL1 84 1000 BSJ-03-123 ADCK3 CABC1 90 1000 BSJ-03-123 ADCK4 ADCK4 91 1000 BSJ-03-123 AKT1 AKT1 100 1000 BSJ-03-123 AKT2 AKT2 98 1000 BSJ-03-123 AKT3 AKT3 100 1000 BSJ-03-123 ALK ALK 100 1000 BSJ-03-123 ALK(C1156Y) ALK 78 1000 BSJ-03-123 ALK(L1196M) ALK 100 1000 BSJ-03-123 AMPK-alpha1 PRKAA1 93 1000 BSJ-03-123 AMPK-alpha2 PRKAA2 100 1000 BSJ-03-123 ANKK1 ANKK1 89 1000 BSJ-03-123 ARK5 NUAK1 98 1000 BSJ-03-123 ASK1 MAP3K5 100 1000 BSJ-03-123 ASK2 MAP3K6 92 1000 BSJ-03-123 AURKA -

Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) JNK-IN-8 AAK1 AAK1 69 1000 JNK-IN-8 ABL1(E255K)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317I)-nonphosphorylated ABL1 87 1000 JNK-IN-8 ABL1(F317I)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317L)-nonphosphorylated ABL1 65 1000 JNK-IN-8 ABL1(F317L)-phosphorylated ABL1 61 1000 JNK-IN-8 ABL1(H396P)-nonphosphorylated ABL1 42 1000 JNK-IN-8 ABL1(H396P)-phosphorylated ABL1 60 1000 JNK-IN-8 ABL1(M351T)-phosphorylated ABL1 81 1000 JNK-IN-8 ABL1(Q252H)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(Q252H)-phosphorylated ABL1 56 1000 JNK-IN-8 ABL1(T315I)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(T315I)-phosphorylated ABL1 92 1000 JNK-IN-8 ABL1(Y253F)-phosphorylated ABL1 71 1000 JNK-IN-8 ABL1-nonphosphorylated ABL1 97 1000 JNK-IN-8 ABL1-phosphorylated ABL1 100 1000 JNK-IN-8 ABL2 ABL2 97 1000 JNK-IN-8 ACVR1 ACVR1 100 1000 JNK-IN-8 ACVR1B ACVR1B 88 1000 JNK-IN-8 ACVR2A ACVR2A 100 1000 JNK-IN-8 ACVR2B ACVR2B 100 1000 JNK-IN-8 ACVRL1 ACVRL1 96 1000 JNK-IN-8 ADCK3 CABC1 100 1000 JNK-IN-8 ADCK4 ADCK4 93 1000 JNK-IN-8 AKT1 AKT1 100 1000 JNK-IN-8 AKT2 AKT2 100 1000 JNK-IN-8 AKT3 AKT3 100 1000 JNK-IN-8 ALK ALK 85 1000 JNK-IN-8 AMPK-alpha1 PRKAA1 100 1000 JNK-IN-8 AMPK-alpha2 PRKAA2 84 1000 JNK-IN-8 ANKK1 ANKK1 75 1000 JNK-IN-8 ARK5 NUAK1 100 1000 JNK-IN-8 ASK1 MAP3K5 100 1000 JNK-IN-8 ASK2 MAP3K6 93 1000 JNK-IN-8 AURKA AURKA 100 1000 JNK-IN-8 AURKA AURKA 84 1000 JNK-IN-8 AURKB AURKB 83 1000 JNK-IN-8 AURKB AURKB 96 1000 JNK-IN-8 AURKC AURKC 95 1000 JNK-IN-8