Phyre 2 Results for Q2FV59
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Protein Expression Profile of Gluconacetobacter Diazotrophicus PAL5, a Sugarcane Endophytic Plant Growth-Promoting Bacterium
Proteomics 2008, 8, 1631–1644 DOI 10.1002/pmic.200700912 1631 RESEARCH ARTICLE Protein expression profile of Gluconacetobacter diazotrophicus PAL5, a sugarcane endophytic plant growth-promoting bacterium Leticia M. S. Lery1, 2, Ana Coelho1, 3, Wanda M. A. von Kruger1, 2, Mayla S. M. Gonc¸alves1, 3, Marise F. Santos1, 4, Richard H. Valente1, 5, Eidy O. Santos1, 3, Surza L. G. Rocha1, 5, Jonas Perales1, 5, Gilberto B. Domont1, 4, Katia R. S. Teixeira1, 6 and Paulo M. Bisch1, 2 1 Rio de Janeiro Proteomics Network, Rio de Janeiro, Brazil 2 Unidade Multidisciplinar de Genômica, Instituto de Biofísica Carlos Chagas Filho, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil 3 Departamento de Genética, Instituto de Biologia, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil 4 Laboratório de Química de Proteínas, Departamento de Bioquímica, Instituto de Química, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil 5 Laboratório de Toxinologia, Departamento de Fisiologia e Farmacodinâmica- Instituto Oswaldo Cruz- Fundac¸ão Oswaldo Cruz, Rio de Janeiro, Rio de Janeiro, Brazil 6 Laboratório de Genética e Bioquímica, Embrapa Agrobiologia, Seropédica, Brazil This is the first broad proteomic description of Gluconacetobacter diazotrophicus, an endophytic Received: September 25, 2007 bacterium, responsible for the major fraction of the atmospheric nitrogen fixed in sugarcane in Revised: December 18, 2007 tropical regions. Proteomic coverage of G. diazotrophicus PAL5 was obtained by two independent Accepted: December 19, 2007 approaches: 2-DE followed by MALDI-TOF or TOF-TOF MS and 1-DE followed by chromatog- raphy in a C18 column online coupled to an ESI-Q-TOF or ESI-IT mass spectrometer. -

Rehmannia Glutinosa-Monocultured Rhizosphere Soil
Comparative Metaproteomic Analysis on Consecutively Rehmannia glutinosa-Monocultured Rhizosphere Soil Linkun Wu1,2, Haibin Wang1,2., Zhixing Zhang1,2., Rui Lin2,3, Zhongyi Zhang1,4, Wenxiong Lin1,2* 1 School of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China, 2 Agroecological Institute, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China, 3 College of Oceanography and Environmental Science, Xiamen University, Xiamen, Fujian, China, 4 Institute of Chinese Medicinal Materials, Henan Agriculture University, Zhengzhou, Henan, China Abstract Background: The consecutive monoculture for most of medicinal plants, such as Rehmannia glutinosa, results in a significant reduction in the yield and quality. There is an urgent need to study for the sustainable development of Chinese herbaceous medicine. Methodology/Principal Findings: Comparative metaproteomics of rhizosphere soil was developed and used to analyze the underlying mechanism of the consecutive monoculture problems of R. glutinosa. The 2D-gel patterns of protein spots for the soil samples showed a strong matrix dependency. Among the spots, 103 spots with high resolution and repeatability were randomly selected and successfully identified by MALDI TOF-TOF MS for a rhizosphere soil metaproteomic profile analysis. These proteins originating from plants and microorganisms play important roles in nutrient cycles and energy flow in rhizospheric soil ecosystem. They function in protein, nucleotide and secondary metabolisms, signal transduction and resistance. Comparative metaproteomics analysis revealed 33 differentially expressed protein spots in rhizosphere soil in response to increasing years of monoculture. Among them, plant proteins related to carbon and nitrogen metabolism and stress response, were mostly up-regulated except a down-regulated protein (glutathione S-transferase) involving detoxification. -
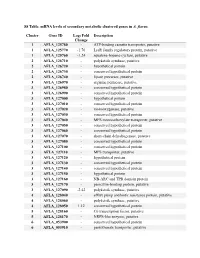
S8 Table. Mrna Levels of Secondary Metabolic Clustered Genes in A
S8 Table. mRNA levels of secondary metabolic clustered genes in A. flavus. Cluster Gene ID Log2 Fold Description Change 1 AFLA_125780 - ATP-binding cassette transporter, putative 1 AFLA_125770 -1.76 LysR family regulatory protein, putative 1 AFLA_125760 -1.24 squalene-hopene-cyclase, putative 2 AFLA_126710 - polyketide synthase, putative 2 AFLA_126720 - hypothetical protein 2 AFLA_126730 - conserved hypothetical protein 2 AFLA_126740 - lipase precursor, putative 3 AFLA_126970 - arginine permease, putative 3 AFLA_126980 - conserved hypothetical protein 3 AFLA_126990 - conserved hypothetical protein 3 AFLA_127000 - hypothetical protein 3 AFLA_127010 - conserved hypothetical protein 3 AFLA_127020 - monooxygenase, putative 3 AFLA_127030 - conserved hypothetical protein 3 AFLA_127040 - MFS monocarboxylate transporter, putative 3 AFLA_127050 - conserved hypothetical protein 3 AFLA_127060 - conserved hypothetical protein 3 AFLA_127070 - short-chain dehydrogenase, putative 3 AFLA_127080 - conserved hypothetical protein 3 AFLA_127100 - conserved hypothetical protein 3 AFLA_127110 - MFS transporter, putative 3 AFLA_127120 - hypothetical protein 3 AFLA_127130 - conserved hypothetical protein 3 AFLA_127140 - conserved hypothetical protein 3 AFLA_127150 - hypothetical protein 3 AFLA_127160 - NB-ARC and TPR domain protein 3 AFLA_127170 - penicillin-binding protein, putative 3 AFLA_127090 -2.42 polyketide synthase, putative 4 AFLA_128040 - efflux pump antibiotic resistance protein, putative 4 AFLA_128060 - polyketide synthase, putative 4 AFLA_128050 -

The Phylogenetic Extent of Metabolic Enzymes and Pathways José Manuel Peregrin-Alvarez, Sophia Tsoka, Christos A
Downloaded from genome.cshlp.org on October 8, 2021 - Published by Cold Spring Harbor Laboratory Press Letter The Phylogenetic Extent of Metabolic Enzymes and Pathways José Manuel Peregrin-Alvarez, Sophia Tsoka, Christos A. Ouzounis1 Computational Genomics Group, The European Bioinformatics Institute, EMBL Cambridge Outstation, Cambridge CB10 1SD, UK The evolution of metabolic enzymes and pathways has been a subject of intense study for more than half a century. Yet, so far, previous studies have focused on a small number of enzyme families or biochemical pathways. Here, we examine the phylogenetic distribution of the full-known metabolic complement of Escherichia coli, using sequence comparison against taxa-specific databases. Half of the metabolic enzymes have homologs in all domains of life, representing families involved in some of the most fundamental cellular processes. We thus show for the first time and in a comprehensive way that metabolism is conserved at the enzyme level. In addition, our analysis suggests that despite the sequence conservation and the extensive phylogenetic distribution of metabolic enzymes, their groupings into biochemical pathways are much more variable than previously thought. One of the fundamental tenets in molecular biology was ex- reliable source of metabolic information. The EcoCyc data- pressed by Monod, in his famous phrase “What is true for base holds information about the full genome and all known Escherichia coli is true for the elephant” (Jacob 1988). For a metabolic pathways of Escherichia coli (Karp et al. 2000). Re- long time, this statement has inspired generations of molecu- cently, the database has been used to represent computational lar biologists, who have used Bacteria as model organisms to predictions of other organisms (Karp 2001). -

WO 2013/180584 Al 5 December 2013 (05.12.2013) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2013/180584 Al 5 December 2013 (05.12.2013) P O P C T (51) International Patent Classification: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, C12N 1/21 (2006.01) C12N 15/74 (2006.01) BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, C12N 15/52 (2006.01) C12P 5/02 (2006.01) DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, C12N 15/63 (2006.01) HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KN, KP, KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, (21) International Application Number: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, PCT/NZ20 13/000095 OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SC, (22) International Filing Date: SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 4 June 2013 (04.06.2013) TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, SZ, TZ, (30) Priority Data: UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, 61/654,412 1 June 2012 (01 .06.2012) US TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, LT, LU, LV, (71) Applicant: LANZATECH NEW ZEALAND LIMITED MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, SM, [NZ/NZ]; 24 Balfour Road, Parnell, Auckland, 1052 (NZ). -

B Number Gene Name Mrna Intensity Mrna Present # of Tryptic
list list sample) short list predicted B number Gene name assignment mRNA present mRNA intensity Gene description Protein detected - Membrane protein detected (total list) detected (long list) membrane sample Proteins detected - detected (short list) # of tryptic peptides # of tryptic peptides # of tryptic peptides # of tryptic peptides # of tryptic peptides Functional category detected (membrane Protein detected - total Protein detected - long b0003 thrB 6781 P 9 P 3 3 P 3 0 homoserine kinase Metabolism of small molecules b0004 thrC 15039 P 18 P 10 P 11 P 10 0 threonine synthase Metabolism of small molecules b0008 talB 20561 P 20 P 13 P 16 P 13 0 transaldolase B Metabolism of small molecules b0009 mog 1296 P 7 0 0 0 0 required for the efficient incorporation of molybdate into molybdoproteins Metabolism of small molecules b0014 dnaK 13283 P 32 P 23 P 24 P 23 0 chaperone Hsp70; DNA biosynthesis; autoregulated heat shock proteins Cell processes b0031 dapB 2348 P 16 P 3 3 P 3 0 dihydrodipicolinate reductase Metabolism of small molecules b0032 carA 9312 P 14 P 8 P 8 P 8 0 carbamoyl-phosphate synthetase, glutamine (small) subunit Metabolism of small molecules b0048 folA 1588 P 7 P 1 2 P 1 0 dihydrofolate reductase type I; trimethoprim resistance Metabolism of small molecules peptidyl-prolyl cis-trans isomerase (PPIase), involved in maturation of outer b0053 surA 3825 P 19 P 4 P 5 P 4 P(m) 1 GenProt membrane proteins (1st module) Cell processes b0054 imp 2737 P 42 P 5 0 0 P(m) 5 GenProt organic solvent tolerance Cell processes b0071 leuD 4770 -

Electric Supporting Information (ESI) Crystal Structure and Functional
Electronic Supplementary Material (ESI) for Chemical Science. This journal is © The Royal Society of Chemistry 2018 Electric Supporting Information (ESI) Crystal structure and functional analysis of large-terpene synthase belonging to a newly found subclass Masahiro Fujihashi,a* Tsutomu Sato,b* Yuma Tanaka,a Daisuke Yamamoto,a Tomoyuki Nishi,b Daijiro Ueda,b Mizuki Murakami,b Yoko Yasuno,c Ai Sekihara,c Kazuma Fuku,c Tetsuro Shinadac & Kunio Mikia* a. Department of Chemistry, Graduate School of Science, Kyoto University, Sakyo-ku, Kyoto 606-8502, Japan. E-mail: [email protected], [email protected] b. Department of Applied Biological Chemistry, Faculty of Agriculture, and Graduate School of Science and Technology, Niigata University, 8050 Ikarashi-2, Niigata 950-2181, Japan. E-mail: [email protected] c. Graduate School of Science, Osaka City University, 3-3-138 Sugimoto, Sumiyoshi, Osaka 558-8585, Japan. S1 Table of Contents Experimental Procedures S3 General procedure S3 Vector construction, expression and purification of BalTS S3 Crystallization and crystallographic analysis of balTS S3 Oligomeric state analysis S4 Analysis of the dimer geometry S4 Vector construction, expression and purification of BsuTS S4 Homology modeling of BsuTS S4 Enzymatic assays S5 Isolation and identification of β-springen S5 Movie Captions S6 Author Contributions S6 Figures Fig. S1 S7 Fig. S2 S8 Fig. S3 S9 Fig. S4 S10 Fig. S5 S17 Fig. S6 S18 Fig. S7, S8 S19 Fig. S9 S20 Fig. S10 S21 Fig. S11 S22 Tables Table S1 S23 Table S2 S24 References S27 S2 Experimental Procedures General procedure NMR spectra were recorded using a Bruker DPX 400 spectrometer at 400 MHz for proton (1H) and 100 MHz for carbon (13C). -

B Number Gene Name Mrna Intensity Mrna
sample) total list predicted B number Gene name assignment mRNA present mRNA intensity Gene description Protein detected - Membrane protein membrane sample detected (total list) Proteins detected - Functional category # of tryptic peptides # of tryptic peptides # of tryptic peptides detected (membrane b0002 thrA 13624 P 39 P 18 P(m) 2 aspartokinase I, homoserine dehydrogenase I Metabolism of small molecules b0003 thrB 6781 P 9 P 3 0 homoserine kinase Metabolism of small molecules b0004 thrC 15039 P 18 P 10 0 threonine synthase Metabolism of small molecules b0008 talB 20561 P 20 P 13 0 transaldolase B Metabolism of small molecules chaperone Hsp70; DNA biosynthesis; autoregulated heat shock b0014 dnaK 13283 P 32 P 23 0 proteins Cell processes b0015 dnaJ 4492 P 13 P 4 P(m) 1 chaperone with DnaK; heat shock protein Cell processes b0029 lytB 1331 P 16 P 2 0 control of stringent response; involved in penicillin tolerance Global functions b0032 carA 9312 P 14 P 8 0 carbamoyl-phosphate synthetase, glutamine (small) subunit Metabolism of small molecules b0033 carB 7656 P 48 P 17 0 carbamoyl-phosphate synthase large subunit Metabolism of small molecules b0048 folA 1588 P 7 P 1 0 dihydrofolate reductase type I; trimethoprim resistance Metabolism of small molecules peptidyl-prolyl cis-trans isomerase (PPIase), involved in maturation of b0053 surA 3825 P 19 P 4 P(m) 1 GenProt outer membrane proteins (1st module) Cell processes b0054 imp 2737 P 42 P 5 P(m) 5 GenProt organic solvent tolerance Cell processes b0071 leuD 4770 P 10 P 9 0 isopropylmalate -

1 AMINO ACIDS Commonly, 21 L-Amino Acids Encoded by DNA Represent the Building Blocks of Animal, Plant, and Microbial Proteins
1 AMINO ACIDS Commonly, 21 L-amino acids encoded by DNA represent the building blocks of animal, plant, and microbial proteins. The basic amino acids encountered in proteins are called proteinogenic amino acids 1.1). Biosynthesis of some of these amino acids proceeds by ribosomal processes only in microorganisms and plants and the ability to synthesize them is lacking in animals, including human beings. These amino acids have to be obtained in the diet (or produced by hydrolysis of body proteins) since they are required for normal good health and are referred to as essential amino acids. The essential amino acids are arginine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, and valine. The rest of encoded amino acids are referred to as non-essential amino acids (alanine, asparagine, aspartic acid, cysteine, glutamic acid, glutamine, glycine, proline, serine, and tyrosine). Arginine and histidine are classified as essential, sometimes as semi-essential amino acids, as their amount synthesized in the body is not sufficient for normal growth of children. Although it is itself non-essential, cysteine (classified as conditionally essential amino acid) can partly replace methionine, which is an essential amino acid. Similarly, tyrosine can partly replace phenylalanine. 1.1 The glutamic acid group 1.1.1 Glutamic acid and glutamine Free ammonium ions are toxic to living cells and are rapidly incorporated into organic compounds. One of such transformations is the reaction of ammonia with 2-oxoglutaric acid from the citric acid cycle to produce L-glutamic acid. This reaction is known as reductive amination. Glutamic acid is accordingly the amino acid generated first as both constituent of proteins and a biosynthetic precursor. -

Terpene Production in the Peel of Sweet Orange Fruits
Genetics and Molecular Biology, 30, 3 (suppl), 841-847 (2007) Copyright by the Brazilian Society of Genetics. Printed in Brazil www.sbg.org.br Research Article Terpene production in the peel of sweet orange fruits Marco A. Takita1,2, Irving J. Berger1, Ana Carolina Basílio-Palmieri1, Kleber M. Borges1, Juliana M. de Souza1 and Maria L.N.P. Targon1 1Centro APTA Citros Sylvio Moreira, Instituto Agronômico de Campinas, Cordeirópolis, SP, Brazil. 2Centro de Pesquisa e Desenvolvimento de Recursos Genéticos Vegetais, Instituto Agronômico de Campinas, Campinas, SP, Brazil. Abstract Terpenoids constitute the largest and most diverse class of natural products. They are important factors for aroma and flavor, and their synthesis is basically done from two compounds: isopentenyl diphosphate and dimethylallyl diphosphate. Isopentenyl diphosphate is synthesized through two different pathways, one that occurs in the cyto- plasm and one in the plastid. With the sequencing of ESTs from citrus, we were able to perform in silico analyses on the pathways that lead to the synthesis of terpenes as well as on the terpene synthases present in sweet orange. Moreover, expression analysis using real-time qPCR was performed to verify the expression pattern of a terpene synthase in plants. The results show that all the pathways for isopentenyl diphosphate are present in citrus and a high expression of terpene synthases seems to have an important role in the constitution of the essential oils of cit- rus. Key words: EST, fruit, terpenoids, orange, essential oil. Received: September 21, 2006; Accepted: July 13, 2007. Introduction ecological, providing defense against herbivores or patho- Citriculture plays a fundamental role in Brazilian ag- gens, attracting animals that disperse pollen and seeds, or ribusiness. -

Developmental Changes in Scots Pine Transcriptome During Heartwood Formation
Developmental changes in Scots pine transcriptome during heartwood formation Kean-Jin Lim, Tanja Paasela, Teemu Teeri University of Helsinki Anni Harju, Martti Venäläinen, Katri Kärkkäinen Luke Punkaharju, June 2016 Heartwood of Scots pine (Pinus sylvestris) is naturally decay resistant Variation in wood extractives correlates with decay resistance — Variation in wood extractives is large and mostly genetic Trees higest in heartwood extractives are most decay resistant. Harju & Venäläinen 2006. Can J. For Res. Heartwood — Does not contain living cells — Reserve materials (e.g. starch) have been converted to ”heartwood substances” — May have different color, lower permeability and increased decay resistance than sapwood — In conifers, heartwood is usually dryer than sapwood Robinia Pinus Picea Heartwood formation Magel 2000 — Type 1 heartwood: Accumulation of (phenolic) extractives takes place in tissue between sapwood and heartwood (transition zone). — Type 2 heartwood: Precursors to phenolics accumulate gradually in ageing sapwood. — Pine heartwood is thought to be of Type 2. Heartwood formation — Much evidence in literature summarizes that heartwood formation takes place during the dormant season. — Pine — From midsummer to autumn (Fukuzawa et al. 1980) — From midsummer to dormant season (Shain and Mackay 1973) — No specific period for heartwood formation (Bergström et al. 1999) Heartwood extractives in Scots pine — Resin acids abietic acid — 50% of heartwood extractives — Stilbenes — 15% of heartwood extractives — Free fatty acids -

Combining Metabolic and Protein Engineering of a Terpenoid Biosynthetic Pathway for Overproduction and Selectivity Control
Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control Effendi Leonarda,1, Parayil Kumaran Ajikumara,1, Kelly Thayerb,c,d,3, Wen-Hai Xiaoa, Jeffrey D. Moa, Bruce Tidorb,c,d, Gregory Stephanopoulosa, and Kristala L. J. Prathera,2 aDepartment of Chemical Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139; bComputer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA 02139; cDepartment of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139; and dDepartment of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA 02139 Edited* by Arnold L. Demain, Drew University, Madison, NJ, and approved June 22, 2010 (received for review May 4, 2010) A common strategy of metabolic engineering is to increase the en- Sequence analysis of these enzymes showed that they are para- dogenous supply of precursor metabolites to improve pathway logous proteins evolved through gene duplications that subse- productivity. The ability to further enhance heterologous produc- quently diverged in functional roles to catalyze the formation tion of a desired compound may be limited by the inherent capacity of different terpenoid structures (16, 17, 19). Particularly, terpe- of the imported pathway to accommodate high precursor supply. noid synthases generate enzyme-bound carbocation intermedi- Here, we present engineered diterpenoid biosynthesis as a case ates that undergo a cascade of rearrangements and quenchings of where insufficient downstream pathway capacity limits high- carbocations to create structural diversity (20). These enzymes level levopimaradiene production in Escherichia coli. To increase are highly promiscuous (21), and the functional promiscuity is levopimaradiene synthesis, we amplified the flux toward isopen- often associated with unwanted product formation and poor cat- tenyl diphosphate and dimethylallyl diphosphate precursors and alytic properties (22).