Identifying Hidden Viable Bacterial Taxa in Tropical Forest Soils Using Amplicon Sequencing of Enrichment Cultures
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Microbiological Study of Fresh White Cheese - 129
Pesic-Mikulec.: Microbiological study of fresh white cheese - 129 - MICROBIOLOGICAL STUDY OF FRESH WHITE CHEESE (A SERBIAN CRAFT VARIETY) D PEŠIĆ-MIKULEC1 − L. JOVANOVIĆ2 e-mail: [email protected] 1 Veterinary Research Institute 11050 Belgrade, Borivoja Stevanovića 2., Serbia and Montenegro 2 BK University, Belgrade, Serbia and Montenegro (Received 7th March 2005, accepted 4th August 2005) Abstract. The levels of several microbial groups of aerobic mesophilic flora, aerobic psychrotrophic flora, lactic acid bacteria, Micrococcaceae, enterococci, Enterobacteriaceae, and molds and yeasts were investigated during the manufacture of fresh white cheese of a Serbian craft variety without the addition of starter culture. This variety of cheese is made in farmhouses from cow, sheep and goat's milk. White fresh cheese from mountain villages of Serbia has economical importance for this area. The study of the microbial characteristics of this cheese constitutes the first step towards the establishment of a starter culture which would allow the making of a product both more uniform and safer. The total microbial counts were high in these variety of cheeses. Almost all the microbial groups reached their maximum counts in curd. Lactic acid bacteria were the major microbial group, reaching count similar to the total aerobic mesophilic flora at all sampling points. Lactococcus lactis subsp. lactis dominated in milk (62,5%) of the isolates obtained in the Man Rogosa Sharpe (MRS) agar at these sampling points, while the Lactobacillus casei subs.casei was the most predominant species (83,5% of isolates obtained at these sampling points). The purpose of this study was to investigate the microflora of white cheeses with special emphasis on the autochthonous lactic acid bacteria involved in fermentation of this cheeses depending on the geographical location where the cheeses were manufactured. -

The Subway Microbiome: Seasonal Dynamics and Direct Comparison Of
Gohli et al. Microbiome (2019) 7:160 https://doi.org/10.1186/s40168-019-0772-9 RESEARCH Open Access The subway microbiome: seasonal dynamics and direct comparison of air and surface bacterial communities Jostein Gohli1* , Kari Oline Bøifot1,2, Line Victoria Moen1, Paulina Pastuszek3, Gunnar Skogan1, Klas I. Udekwu4 and Marius Dybwad1,2 Abstract Background: Mass transit environments, such as subways, are uniquely important for transmission of microbes among humans and built environments, and for their ability to spread pathogens and impact large numbers of people. In order to gain a deeper understanding of microbiome dynamics in subways, we must identify variables that affect microbial composition and those microorganisms that are unique to specific habitats. Methods: We performed high-throughput 16S rRNA gene sequencing of air and surface samples from 16 subway stations in Oslo, Norway, across all four seasons. Distinguishing features across seasons and between air and surface were identified using random forest classification analyses, followed by in-depth diversity analyses. Results: There were significant differences between the air and surface bacterial communities, and across seasons. Highly abundant groups were generally ubiquitous; however, a large number of taxa with low prevalence and abundance were exclusively present in only one sample matrix or one season. Among the highly abundant families and genera, we found that some were uniquely so in air samples. In surface samples, all highly abundant groups were also well represented in air samples. This is congruent with a pattern observed for the entire dataset, namely that air samples had significantly higher within-sample diversity. We also observed a seasonal pattern: diversity was higher during spring and summer. -

Dissertation Implementing Organic Amendments To
DISSERTATION IMPLEMENTING ORGANIC AMENDMENTS TO ENHANCE MAIZE YIELD, SOIL MOISTURE, AND MICROBIAL NUTRIENT CYCLING IN TEMPERATE AGRICULTURE Submitted by Erika J. Foster Graduate Degree Program in Ecology In partial fulfillment of the requirements For the Degree of Doctor of Philosophy Colorado State University Fort Collins, Colorado Summer 2018 Doctoral Committee: Advisor: M. Francesca Cotrufo Louise Comas Charles Rhoades Matthew D. Wallenstein Copyright by Erika J. Foster 2018 All Rights Reserved i ABSTRACT IMPLEMENTING ORGANIC AMENDMENTS TO ENHANCE MAIZE YIELD, SOIL MOISTURE, AND MICROBIAL NUTRIENT CYCLING IN TEMPERATE AGRICULTURE To sustain agricultural production into the future, management should enhance natural biogeochemical cycling within the soil. Strategies to increase yield while reducing chemical fertilizer inputs and irrigation require robust research and development before widespread implementation. Current innovations in crop production use amendments such as manure and biochar charcoal to increase soil organic matter and improve soil structure, water, and nutrient content. Organic amendments also provide substrate and habitat for soil microorganisms that can play a key role cycling nutrients, improving nutrient availability for crops. Additional plant growth promoting bacteria can be incorporated into the soil as inocula to enhance soil nutrient cycling through mechanisms like phosphorus solubilization. Since microbial inoculation is highly effective under drought conditions, this technique pairs well in agricultural systems using limited irrigation to save water, particularly in semi-arid regions where climate change and population growth exacerbate water scarcity. The research in this dissertation examines synergistic techniques to reduce irrigation inputs, while building soil organic matter, and promoting natural microbial function to increase crop available nutrients. The research was conducted on conventional irrigated maize systems at the Agricultural Research Development and Education Center north of Fort Collins, CO. -

Product List 2008/09
Product List 2008/09 Part of Thermo Fisher Scientific Oxoid Chromogenic Media … Sheer Brilliance™ to reflect this, the names of products are being changed so that the current range now includes: Brilliance Bacillus cereus Agar Brilliance Candida Agar Brilliance E. coli/coliform Agar Brilliance E. coli/coliform Selective Agar Brilliance Enterobacter sakazakii Agar (DFI) Brilliance Listeria Agar Brilliance UTI Agar Brilliance UTI Clarity Agar Brilliance Salmonella Agar The chromogens used in our Brilliance range of chromogenic media ensure that colony colours are vivid, allowing rapid, easy differentiation and presumptive identification of the organism in question. So remember Brilliance – chromogenic media that delivers clearly visible answers on a single culture plate. Contents 1 Culture Media & Supplements 2 Supplementary Reagents 20 Antibiotic Single Supplements 21 Biochemical Reagents 22 Laboratory Preparations & Biological Extracts 22 Veggietones 24 Bagged Media 25 Dip Slides 25 Atmosphere Generation 26 AGS 26 Traditional Systems 27 Food Testing 28 DuPont Qualicon 29 BAX® System, Test and Components 29 Culture Media for the BAX System 30 RiboPrinter® Microbial Characterisation System 31 Lateral Flow System Range 31 Blood Culture 32 Signal 32 Wampole Isolator® 32 Antimicrobial Susceptibility Testing 33 M.I.C.Evaluator Strips 33 aura image 34 AST Accessories 34 Antimicrobial Susceptibility Testing Discs 35 Miscellaneous 38 Sundries 38 Publications 38 Diagnostics 39 Biochemical I.D. 39 Toxin Detection Kits 40 Agglutination Tests 41 Enzyme Immunoassays 42 Immunofluorescence Assays 43 Lateral Flow Assays 44 Environmental Monitoring and Process Simulation 45 Oxoid Air Sampler 45 Environmental Monitoring Swabs 45 Prepared Media 45 Dehydrated Media 45 Process Simulation 46 2 Culture Media and Supplements A wide range of general purpose, enrichment, selective and differential Anaerobe Basal Broth media, diluents and supplements a medium for general growth of anaerobes NB: information provided here is intended only as an aid to purchasing. -

Microbiological and Metagenomic Characterization of a Retail Delicatessen Galotyri-Like Fresh Acid-Curd Cheese Product
fermentation Article Microbiological and Metagenomic Characterization of a Retail Delicatessen Galotyri-Like Fresh Acid-Curd Cheese Product John Samelis 1,* , Agapi I. Doulgeraki 2,* , Vasiliki Bikouli 2, Dimitrios Pappas 3 and Athanasia Kakouri 1 1 Dairy Research Department, Hellenic Agricultural Organization ‘DIMITRA’, Katsikas, 45221 Ioannina, Greece; [email protected] 2 Hellenic Agricultural Organization ‘DIMITRA’, Institute of Technology of Agricultural Products, 14123 Lycovrissi, Greece; [email protected] 3 Skarfi EPE—Pappas Bros Traditional Dairy, 48200 Filippiada, Greece; [email protected] * Correspondence: [email protected] (J.S.); [email protected] (A.I.D.); Tel.: +30-2651094789 (J.S.); +30-2102845940 (A.I.D.) Abstract: This study evaluated the microbial quality, safety, and ecology of a retail delicatessen Galotyri-like fresh acid-curd cheese traditionally produced by mixing fresh natural Greek yogurt with ‘Myzithrenio’, a naturally fermented and ripened whey cheese variety. Five retail cheese batches (mean pH 4.1) were analyzed for total and selective microbial counts, and 150 presumptive isolates of lactic acid bacteria (LAB) were characterized biochemically. Additionally, the most and the least diversified batches were subjected to a culture-independent 16S rRNA gene sequencing analysis. LAB prevailed in all cheeses followed by yeasts. Enterobacteria, pseudomonads, and staphylococci were present as <100 viable cells/g of cheese. The yogurt starters Streptococcus thermophilus and Lactobacillus delbrueckii were the most abundant LAB isolates, followed by nonstarter strains of Lactiplantibacillus, Lacticaseibacillus, Enterococcus faecium, E. faecalis, and Leuconostoc mesenteroides, Citation: Samelis, J.; Doulgeraki, A.I.; whose isolation frequency was batch-dependent. Lactococcus lactis isolates were sporadic, except Bikouli, V.; Pappas, D.; Kakouri, A. Microbiological and Metagenomic for one cheese batch. -

Food Microbiology
Food Microbiology Food Water Dairy Beverage Online Ordering Available Food, Water, Dairy, & Beverage Microbiology Table of Contents 1 Environmental Monitoring Contact Plates 3 Petri Plates 3 Culture Media for Air Sampling 4 Environmental Sampling Boot Swabs 6 Environmental Testing Swabs 8 Surface Sanitizers 8 Hand Sanitation 9 Sample Preparation - Dilution Vials 10 Compact Dry™ 12 HardyCHROM™ Chromogenic Culture Media 15 Prepared Media 24 Agar Plates for Membrane Filtration 26 CRITERION™ Dehydrated Culture Media 28 Pathogen Detection Environmental With Monitoring Contact Plates Baird Parker Agar Friction Lid For the selective isolation and enumeration of coagulase-positive staphylococci (Staphylococcus aureus) on environmental surfaces. HardyCHROM™ ECC 15x60mm contact plate, A chromogenic medium for the detection, 10/pk ................................................................................ 89407-364 differentiation, and enumeration of Escherichia coli and other coliforms from environmental surfaces (E. coli D/E Neutralizing Agar turns blue, coliforms turn red). For the enumeration of environmental organisms. 15x60mm plate contact plate, The media is able to neutralize most antiseptics 10/pk ................................................................................ 89407-354 and disinfectants that may inhibit the growth of environmental organisms. Malt Extract 15x60mm contact plate, Malt Extract is recommended for the cultivation and 10/pk ................................................................................89407-482 -

View Our Full Water Sampling Vials Product Offering
TABLE OF CONTENTS Environmental Monitoring 1 Sample Prep and Dilution 8 Dehydrated Culture Media - Criterion™ 12 Prepared Culture Media 14 Chromogenic Media - HardyCHROM™ 18 CompactDry™ 20 Quality Control 24 Membrane Filtration 25 Rapid Tests 26 Lab Supplies/Sample Collection 27 Made in the USA Headquarters Distribution Centers 1430 West McCoy Lane Santa Maria, California Santa Maria, CA 93455 Olympia, Washington 800.266.2222 : phone Salt Lake City, Utah [email protected] Phoenix, Arizona HardyDiagnostics.com Dallas, Texas Springboro, Ohio Hardy Diagnostics has a Quality Lake City, Florida Management System that is certified Albany, New York to ISO 13485 and is a FDA licensed Copyright © 2020 Hardy Diagnostics Raleigh, North Carolina medical device manufacturer. Environmental Monitoring Hardy Diagnostics offers a wide selection of quality products intended to help keep you up to date with regulatory compliance, ensure the efficacy of your cleaning protocol, and properly monitor your environment. 800.266.2222 [email protected] HardyDiagnostics.com 1 Environmental Monitoring Air Sampling TRIO.BAS™ Impact Air Samplers introduced the newest generation of microbial air sampling. These ergonomically designed instruments combine precise air sampling with modern connectivity to help you properly assess the air quality of your laboratory and simplify your process. MONO DUO Each kit includes: Each kit includes: TRIO.BAS™ MONO air sampler, induction TRIO.BAS™ DUO air sampler, battery battery charger and cable, aspirating charger -

WHO Global Foodborne Infections Network
WHO Global Foodborne Infections Network (formerly WHO Global Salm-Surv) "A WHO network building capacity to detect, control and prevent foodborne and other enteric infections from farm to table” Laboratory Protocol “Isolation of Salmonella spp. From Food and Animal Faeces ” 5th Ed. June 2010 1 IMPORTANT NOTES: 1) This procedure is based on the ISO protocol: 6579:2002 “Microbiology of food and animal feeding stuffs -- Horizontal method for the detection of Salmonella spp.”4. This protocol is intended to provide guidance for the testing of suspect food items/ animal faecal specimens identified via foodborne disease surveillance programmes. Regulatory agencies (Ministries of Health, Agriculture, Commerce, etc) have specific testing requirements, different from this protocol, which much be used to test samples collected for regulatory testing (example: import/export or product recall). Prior to performing any official, legal, or regulatory testing, the reader should confirm the appropriate protocol through consult with in-country regulatory authorities. 2) This protocol is intended only to be used on food samples and animal faeces. This protocol should not be used for the testing of human faeces. Foreword: Infections due to Salmonella spp. remain a global problem. These infections may cause significant morbidity and mortality both in humans and production animals as well as considerable economic losses. Salmonella spp. are typically transmitted among humans and animals via a fecal-oral route, usually through the consumption of contaminated food or water. Timely identification and serotyping of Salmonella from clinical specimens facilitates outbreak detection and patient management while prompt and accurate detection of Salmonella spp. in contaminated food or water provides an opportunity to prevent the contaminated food from entering the food supply. -
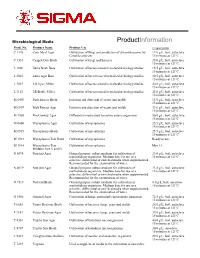
Nutrient Broth (N7519)
Microbiological Media ProductInformation Prod. No. Product Name Product Use Preparation C 1176 Corn Meal Agar Cultivation of fungi and production of chlamydospores by 17.0 g/L, boil, autoclave Candida albicans 15 minutes at 121ºC. C 1551 Czapek Dox Broth Cultivation of fungi and bacteria 35.0 g/L, boil, autoclave 15 minutes at 121ºC. L 1900 Luria Broth Base Cultivation of bacteria used in molecular biology studies. 15.5 g/L, boil, autoclave 15 minutes at 121ºC. L 2025 Luria Agar Base Cultivation of bacteria used in molecular biology studies. 30.5 g/L, boil, autoclave 15 minutes at 121ºC. L 3027 LB Agar, Miller Cultivation of bacteria used in molecular biology studies. 40.0 g/L; boil, autoclave 15 minutes at 121ºC. L 3152 LB Broth, Miller Cultivation of bacteria used in molecular biology studies. 25.0 g/L; boil, autoclave 15 minutes at 121ºC. M 6409 Malt Extract Broth Isolation and detection of yeasts and molds 15.0 g/L, boil, autoclave 15 minutes at 121ºC. M 6907 Malt Extract Agar Isolation and detection of yeasts and molds 33.6 g/L, boil, autoclave 15 minutes at 121ºC. M 7408 MacConkey Agar Differential media used to isolate enteric organisms 50.0 g/L; boil, autoclave 15 minutes at 121ºC M 0660 Mycoplasma Agar Cultivation of mycoplasma 35.5 g/L; boil, autoclave 15 minutes at 121ºC M 0535 Mycoplasma Broth Cultivation of mycoplasma 25.5 g/L; boil, autoclave 15 minutes at 121ºC M 1539 Mycoplasma Test Broth Cultivation of mycoplasma Ready to use. M 1914 Mycoplasma Test Cultivation of mycoplasma Mix 1:1. -

Compile.Xlsx
Silva OTU GS1A % PS1B % Taxonomy_Silva_132 otu0001 0 0 2 0.05 Bacteria;Acidobacteria;Acidobacteria_un;Acidobacteria_un;Acidobacteria_un;Acidobacteria_un; otu0002 0 0 1 0.02 Bacteria;Acidobacteria;Acidobacteriia;Solibacterales;Solibacteraceae_(Subgroup_3);PAUC26f; otu0003 49 0.82 5 0.12 Bacteria;Acidobacteria;Aminicenantia;Aminicenantales;Aminicenantales_fa;Aminicenantales_ge; otu0004 1 0.02 7 0.17 Bacteria;Acidobacteria;AT-s3-28;AT-s3-28_or;AT-s3-28_fa;AT-s3-28_ge; otu0005 1 0.02 0 0 Bacteria;Acidobacteria;Blastocatellia_(Subgroup_4);Blastocatellales;Blastocatellaceae;Blastocatella; otu0006 0 0 2 0.05 Bacteria;Acidobacteria;Holophagae;Subgroup_7;Subgroup_7_fa;Subgroup_7_ge; otu0007 1 0.02 0 0 Bacteria;Acidobacteria;ODP1230B23.02;ODP1230B23.02_or;ODP1230B23.02_fa;ODP1230B23.02_ge; otu0008 1 0.02 15 0.36 Bacteria;Acidobacteria;Subgroup_17;Subgroup_17_or;Subgroup_17_fa;Subgroup_17_ge; otu0009 9 0.15 41 0.99 Bacteria;Acidobacteria;Subgroup_21;Subgroup_21_or;Subgroup_21_fa;Subgroup_21_ge; otu0010 5 0.08 50 1.21 Bacteria;Acidobacteria;Subgroup_22;Subgroup_22_or;Subgroup_22_fa;Subgroup_22_ge; otu0011 2 0.03 11 0.27 Bacteria;Acidobacteria;Subgroup_26;Subgroup_26_or;Subgroup_26_fa;Subgroup_26_ge; otu0012 0 0 1 0.02 Bacteria;Acidobacteria;Subgroup_5;Subgroup_5_or;Subgroup_5_fa;Subgroup_5_ge; otu0013 1 0.02 13 0.32 Bacteria;Acidobacteria;Subgroup_6;Subgroup_6_or;Subgroup_6_fa;Subgroup_6_ge; otu0014 0 0 1 0.02 Bacteria;Acidobacteria;Subgroup_6;Subgroup_6_un;Subgroup_6_un;Subgroup_6_un; otu0015 8 0.13 30 0.73 Bacteria;Acidobacteria;Subgroup_9;Subgroup_9_or;Subgroup_9_fa;Subgroup_9_ge; -

Bacterial Cultures Care Guide SCIENTIFIC Introduction Use the Following Guide to Learn How to Properly Care for Bacterial Cultures
Bacterial Cultures Care Guide SCIENTIFIC Introduction Use the following guide to learn how to properly care for bacterial cultures. BIO FAX! Safety Precautions After use, agar plates will likely contain viable microbes. Although the bacteria are not likely to be pathogenic, do not open the plates unnecessarily. Use sterile techniques at all times when handling bacterial cultures. When plates are done being used, they should be autoclaved or opened under a 10% bleach solution and soaked for at least one hour. Bleach solution is a corrosive liquid that may dis- color clothing and cause skin burns. Avoid contact of bleach with heat, acids and organic materials; chlorine gas will be generated. Wear chemical splash goggles, chemical-resistant gloves, and a chemical-resistant apron. Wash hands thoroughly with soap and water before leaving the laboratory. Please follow all laboratory safety guidelines. Culturing and Maintenance Prior to receiving bacterial cultures it is helpful to be aware of the required conditions for the particular strain of bacteria purchased. Each species has specific conditions that are necessary for optimal growth. Refer to the Bacterial Cultures Table below for incubation temperature and the appropriate general medium. Bacterial Cultures Table Description (Genus species) Incubation Temperature† Medium Bacillus cereus (20–35) 30 °C Nutrient agar Bacillus mycoides 30 °C Nutrient agar Bacillus megaterium (25–35) 30 °C Nutrient agar Bacillus subtilis (25–35) 25–30 °C Nutrient agar Enterobacter aerogenes (30–37) 30 °C Nutrient -

Inter-Domain Horizontal Gene Transfer of Nickel-Binding Superoxide Dismutase 2 Kevin M
bioRxiv preprint doi: https://doi.org/10.1101/2021.01.12.426412; this version posted January 13, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Inter-domain Horizontal Gene Transfer of Nickel-binding Superoxide Dismutase 2 Kevin M. Sutherland1,*, Lewis M. Ward1, Chloé-Rose Colombero1, David T. Johnston1 3 4 1Department of Earth and Planetary Science, Harvard University, Cambridge, MA 02138 5 *Correspondence to KMS: [email protected] 6 7 Abstract 8 The ability of aerobic microorganisms to regulate internal and external concentrations of the 9 reactive oxygen species (ROS) superoxide directly influences the health and viability of cells. 10 Superoxide dismutases (SODs) are the primary regulatory enzymes that are used by 11 microorganisms to degrade superoxide. SOD is not one, but three separate, non-homologous 12 enzymes that perform the same function. Thus, the evolutionary history of genes encoding for 13 different SOD enzymes is one of convergent evolution, which reflects environmental selection 14 brought about by an oxygenated atmosphere, changes in metal availability, and opportunistic 15 horizontal gene transfer (HGT). In this study we examine the phylogenetic history of the protein 16 sequence encoding for the nickel-binding metalloform of the SOD enzyme (SodN). A comparison 17 of organismal and SodN protein phylogenetic trees reveals several instances of HGT, including 18 multiple inter-domain transfers of the sodN gene from the bacterial domain to the archaeal domain.