Divergence of TERMINAL FLOWER1-Like Genes in Rosaceae
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Stomata Size in Relation to Ploidy Level in North American Hawthorns (Crataegus, Rosaceae) Author(S): Brechann V
Stomata Size in Relation to Ploidy Level in North American Hawthorns (Crataegus, Rosaceae) Author(s): Brechann V. McGoey Kelvin Chau Timothy A. Dickinson Source: Madroño, 61(2):177-193. 2014. Published By: California Botanical Society DOI: http://dx.doi.org/10.3120/0024-9637-61.2.177 URL: http://www.bioone.org/doi/full/10.3120/0024-9637-61.2.177 BioOne (www.bioone.org) is a nonprofit, online aggregation of core research in the biological, ecological, and environmental sciences. BioOne provides a sustainable online platform for over 170 journals and books published by nonprofit societies, associations, museums, institutions, and presses. Your use of this PDF, the BioOne Web site, and all posted and associated content indicates your acceptance of BioOne’s Terms of Use, available at www.bioone.org/page/ terms_of_use. Usage of BioOne content is strictly limited to personal, educational, and non-commercial use. Commercial inquiries or rights and permissions requests should be directed to the individual publisher as copyright holder. BioOne sees sustainable scholarly publishing as an inherently collaborative enterprise connecting authors, nonprofit publishers, academic institutions, research libraries, and research funders in the common goal of maximizing access to critical research. MADRON˜ O, Vol. 61, No. 2, pp. 177–193, 2014 STOMATA SIZE IN RELATION TO PLOIDY LEVEL IN NORTH AMERICAN HAWTHORNS (CRATAEGUS,ROSACEAE) BRECHANN V. MCGOEY Department of Ecology and Evolutionary Biology, University of Toronto, Toronto, ON, Canada M5S 3B2 [email protected] KELVIN CHAU Canadian Food Inspection Agency, 1124 Finch Ave. W, Unit 2, Toronto, ON, Canada M3J 2E2 TIMOTHY A. DICKINSON Green Plant Herbarium (TRT), Department of Natural History, Royal Ontario Museum, 100 Queen’s Park, Toronto, ON, Canada M5S 2C6, and Department of Ecology and Evolutionary Biology, University of Toronto, Toronto, ON, Canada M5S 3B2 ABSTRACT The impacts of ploidy level changes on plant physiology and ecology present interesting avenues of research, and many questions remain unanswered. -

A Preliminary List of the Flora of the Perkiomen Region Whorten A
Ursinus College Digital Commons @ Ursinus College Faculty Monographs and Chapters 1924 A Preliminary List of the Flora of the Perkiomen Region Whorten A. Kline Ursinus College Thomas R. Brendle Joseph R. Mumbauer Follow this and additional works at: https://digitalcommons.ursinus.edu/faculty_books Part of the Botany Commons, and the Environmental Studies Commons Click here to let us know how access to this document benefits oy u. Recommended Citation Kline, Whorten A.; Brendle, Thomas R.; and Mumbauer, Joseph R., "A Preliminary List of the Flora of the Perkiomen Region" (1924). Faculty Monographs and Chapters. 11. https://digitalcommons.ursinus.edu/faculty_books/11 This Book is brought to you for free and open access by Digital Commons @ Ursinus College. It has been accepted for inclusion in Faculty Monographs and Chapters by an authorized administrator of Digital Commons @ Ursinus College. For more information, please contact [email protected]. THE FLORA OF The Perkiomen Region BASED ON THE FIELD WORK OF PROF. DR.SSHftvA. KLINE \U tUtkMj T. ROYCE BRENDLE JOSEPH R. mUMBAUER COMPILED BY THE ABOVE 1924 FOREWORD Nobody appreciates more than we the incompleteness and im perfections of this preliminary list of the flora of the Perkiomen Region. W'e hesitated to have it published. We trust that its failings may lead to the publication of a "Flora of the Perk~omen Region." The first botanist of the valley must have been Muhlenberg. It is not known that he wrote anything pertaining particularly to our region. For many years members of the Philadelphia Botanical Club have collected in the valley. Many of their specimens are in the herbarium at the Academy of Natural Sciences, Philadelphia. -

Natural Communities of Michigan: Classification and Description
Natural Communities of Michigan: Classification and Description Prepared by: Michael A. Kost, Dennis A. Albert, Joshua G. Cohen, Bradford S. Slaughter, Rebecca K. Schillo, Christopher R. Weber, and Kim A. Chapman Michigan Natural Features Inventory P.O. Box 13036 Lansing, MI 48901-3036 For: Michigan Department of Natural Resources Wildlife Division and Forest, Mineral and Fire Management Division September 30, 2007 Report Number 2007-21 Version 1.2 Last Updated: July 9, 2010 Suggested Citation: Kost, M.A., D.A. Albert, J.G. Cohen, B.S. Slaughter, R.K. Schillo, C.R. Weber, and K.A. Chapman. 2007. Natural Communities of Michigan: Classification and Description. Michigan Natural Features Inventory, Report Number 2007-21, Lansing, MI. 314 pp. Copyright 2007 Michigan State University Board of Trustees. Michigan State University Extension programs and materials are open to all without regard to race, color, national origin, gender, religion, age, disability, political beliefs, sexual orientation, marital status or family status. Cover photos: Top left, Dry Sand Prairie at Indian Lake, Newaygo County (M. Kost); top right, Limestone Bedrock Lakeshore, Summer Island, Delta County (J. Cohen); lower left, Muskeg, Luce County (J. Cohen); and lower right, Mesic Northern Forest as a matrix natural community, Porcupine Mountains Wilderness State Park, Ontonagon County (M. Kost). Acknowledgements We thank the Michigan Department of Natural Resources Wildlife Division and Forest, Mineral, and Fire Management Division for funding this effort to classify and describe the natural communities of Michigan. This work relied heavily on data collected by many present and former Michigan Natural Features Inventory (MNFI) field scientists and collaborators, including members of the Michigan Natural Areas Council. -

Plant Descriptions 2018 4/22/2018
Tyler Plant Sale - Plant Descriptions 2018 4/22/2018 TypeDesc Botanical Common Season of Exposure Size Description Name Name Interest Woody: Vine Clematis Clematis Summer to Sun to 8-10' Clematis 'Cardinal Wyszynski' dazzles your garden with huge 8" glowing 'Cardinal Fall Partial crimson flowers. The vibrant flowers are accented with darker crimson Wyszynski' Shade anthers and light pink filaments. Blooms in June-July and again in September. Attracts pollinators. Easy to grow in a rich, porous, alkaline soil. Provide shade for the roots with a generous layer of mulch or a shallow-rooted groundcover near the base of the vine. Received the Golden Medal at 'Plantarium' in 1990. Woody: Vine Clematis Hybrid Summer Sun to 6-8’ Fully double white flowers have yellow anthers and green outer petals. 'Duchess of Clematis Partial They are borne on the previous year’s growth and the current season’s Edinburgh' Shade new growth. This clematis does not require heavy pruning, remove only weak or dead stems in late spring. Tolerates most garden soils, needs protection from cold winds. Woody: Vine Clematis Clematis Early Sun to 8-10’ A beautiful, compact vine that covers itself with 5” shell pink flowers in 'Hagley Summer Partial summer. 'Hagley Hybrid' is also know as Pink Chiffon. This is a large- Hybrid' Shade flowering clematis that can be grown as a container plant. It is best keep out of full sun to prevent bleaching of flowers. Prefers moist, well-drained soil and for best results, mulch. TypeDesc Botanical Common Season of Exposure Size Description Name Name Interest Woody: Vine Clematis x Clematis Summer to Sun to 6-10' This deciduous hybrid clematis, has unusual and very striking deep blue durandii Fall Partial flowers with creamy stamens on a non-clinging, scrambling vine. -

Filipendula Ulmaria (L.) Maxim
6 May 2020 EMA/HMPC/595722/2019 Committee on Herbal Medicinal Products (HMPC) Addendum to Assessment report on Filipendula ulmaria (L.) Maxim. (= Spiraea ulmaria L.), herba Rapporteur(s) B Kroes Assessor(s) Jan van der Nat Peer-reviewer J Wiesner HMPC decision on review of monograph Filipendula ulmaria (L.) Maxim. (= Spiraea 30 January 2018 ulmaria L.), herba adopted on July 2011 Call for scientific data (start and end date) From 30 April 2018 to 31 July 2018 Adoption by Committee on Herbal Medicinal 6 May 2020 Products (HMPC) Review of new data on Filipendula ulmaria (L.) Maxim., herba Periodic review (from 2011 to 2018) Scientific data (e.g. non-clinical and clinical safety data, clinical efficacy data) Pharmacovigilance data (e.g. data from EudraVigilance, VigiBase, national databases) Scientific/Medical/Toxicological databases: Scopus, PubMed, Embase, ToxNet Other Regulatory practice Old market overview in AR (i.e. products fulfilling 30/15 years on the market) New market overview (including pharmacovigilance actions taken in member states) – information from Member States (reporting between November 2018 and January 2019): Official address Domenico Scarlattilaan 6 ● 1083 HS Amsterdam ● The Netherlands Address for visits and deliveries Refer to www.ema.europa.eu/how-to-find-us Send us a question Go to www.ema.europa.eu/contact Telephone +31 (0)88 781 6000 An agency of the European Union © European Medicines Agency, 2020. Reproduction is authorised provided the source is acknowledged. Referral Ph.Eur. monograph: Filipendulae ulmariae herba 04/2013:1868 Currently: request for revision: replacement of hexane in TLC identification. Other Consistency (e.g. scientific decisions taken by HMPC) Public statements or other decisions taken by HMPC Consistency with other monographs within the therapeutic area Other Availability of new information (i.e. -

Original Research Article
1 Original Research Article 2 3 THE MALOIDEAE (ROSACEAE) STRUCTURAL AND FUNCTIONAL FEATURES 4 DETERMINING PASSIVE IMMUNITY TO MYCOSIS 5 6 7 With the help of microscopic methods the leaves and fruits surface tissues of plants of four 8 genera of the Maloideae subfamily were screened: Malus Mill., Pyrus L., Cydonia Mill., 9 Mespilus L., as model objects, and attempts were made to explain the dependence of mycosis 10 damage on microstructural features. The species composition of fungi that cause damage to the 11 Maloideae leaves and fruits in the Russia southern regions is analyzed. It is established that 12 among pathogens with different types of parasitism there are common excitants, as well as 13 highly specialized, more represented on Mespilus germanica. Higher resistance to the complex 14 of fungal diseases, in comparison with apple and pear, was found in quince and medlar. This 15 stability at the initial stage of the pathological process is associated with structural features such 16 as micromorphology of the fruits and stomata cuticle in the abaxial leaves epidermis. The leaves 17 stomatal cracks of the medlar are narrow with raised outgrowths, on the surface of the fruit – the 18 layered structure of the cuticular layer. Quince has a powerful continuous cuticular cover. 19 Compared with Malus and Pyrus, Cydonia and Mespilus also have a large (30 % or more) 20 polyphenol content in the pericarp outer layer cells. In addition to the gender-specific differences 21 in the microstructure of the integumentary tissues and the content of polyphenols affecting the 22 resistance to pathogens at the stage of their penetration, general patterns of leaf surface 23 formation, such as hypostomacy, anomocytic stomata, folded microrelief of the cuticular surface, 24 and the presence of single and multicellular trichomes are noted. -

Production, Pomological and Nutraceutical Properties of Apricot
1 Production, pomological and nutraceutical properties of apricot Khaled Moustafa1* and Joanna Cross2 1Editor of ArabiXiv (arabixiv.org), Paris, France 2Nigde Omer Halisdemir University, Nigde, Turkey Correspondence: [email protected] Abstract Apricot (Prunus sp.) is an important fruit crop worldwide. Despite recent advances in apricot research, much is still to be done to improve its productivity and environmental adaptability. The availability of wild apricot germplasms with economically interesting traits is a strong incentive to increase research panels toward improving its economic, environmental and nutritional characteristics. New technologies and genomic studies have generated a large amount of raw data that the mining and exploitation can help decrypt the biology of apricot and enhance its agronomic values. Here, we outline recent findings in relation to apricot production, pomological and nutraceutical properties. In particular, we retrace its origin from central Asia and the path it took to attain Europe and other production areas around the Mediterranean basin while locating it in the rosaceae family and referring to its genetic diversities and new attempts of classification. The production, nutritional, and nutraceutical importance of apricot are recapped in an easy readable and comparable way. We also highlight and discuss the effects of late frost damages on apricot production over different growth stages, from swollen buds to green fruits formation. Issues related to the length of production season and biotic and abiotic environmental challenges are also discussed with future perspective on how to lengthen the production season without compromising the fruit quality and productivity. Keywords Apricot kernel oil, plum pox virus, prunus armeniaca, spring frost, stone fruit, sharka. -

South Carolina Wildflowers by Color and Season
SOUTH CAROLINA WILDFLOWERS *Chokeberry (Aronia arbutifolia) Silky Camellia (Stewartia malacodendron) BY COLOR AND SEASON Mountain Camelia (Stewartia ovata) Dwarf Witch Alder (Fothergilla gardenii) Revised 10/2007 by Mike Creel *Wild Plums (Prunus angustifolia, americana) 155 Cannon Trail Road Flatwoods Plum (Prunus umbellata) Lexington, SC 29073 *Shadberry or Sarvis Tree (Amelanchier arborea, obovata) Phone: (803) 359-2717 E-mail: [email protected] Fringe Tree (Chionanthus virginicus) Yellowwood Tree (Cladratis kentuckeana) Silverbell Tree (Halesia carolina, etc.) IDENTIFY PLANTS BY COLOR, THEN Evergreen Cherry Laurel (Prunus caroliniana) SEASON . Common ones in bold print. Hawthorn (Crataegus viridis, marshalli, etc.) Storax (Styrax americana, grandifolia) Wild Crabapple (Malus angustifolia) WHITE Wild Cherry (Prunus serotina) SPRING WHITE Dec. 1 to May 15 SUMMER WHITE May 15 to Aug. 7 *Atamasco Lily (Zephyranthes atamasco) *Swamp Spiderlily (Hymenocallis crassifolia) Carolina Anemone (Anemone caroliniana) Rocky Shoals Spiderlily (Hymenocallis coronaria) Lance-leaved Anemone (Anemone lancifolia) Colic Root (Aletris farinosa) Meadow Anemone (Anemone canadensis) Fly-Poison (Amianthium muscaetoxicum) American Wood Anemone (Anemone quinquefolia) Angelica (Angelica venosa) Wild Indigo (Baptisia bracteata) Ground Nut Vine (Apios americana) Sandwort (Arenaria caroliniana) Indian Hemp (Apocynum cannabium) American Bugbane (Cimicifuga americana) Sand Milkweed (Asclepias humistrata) Cohosh Bugbane (Cimicifuga racemosa) White Milkweed (Asclepias -

Phylogeny of Maleae (Rosaceae) Based on Multiple Chloroplast Regions: Implications to Genera Circumscription
Hindawi BioMed Research International Volume 2018, Article ID 7627191, 10 pages https://doi.org/10.1155/2018/7627191 Research Article Phylogeny of Maleae (Rosaceae) Based on Multiple Chloroplast Regions: Implications to Genera Circumscription Jiahui Sun ,1,2 Shuo Shi ,1,2,3 Jinlu Li,1,4 Jing Yu,1 Ling Wang,4 Xueying Yang,5 Ling Guo ,6 and Shiliang Zhou 1,2 1 State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China 2University of the Chinese Academy of Sciences, Beijing 100043, China 3College of Life Science, Hebei Normal University, Shijiazhuang 050024, China 4Te Department of Landscape Architecture, Northeast Forestry University, Harbin 150040, China 5Key Laboratory of Forensic Genetics, Institute of Forensic Science, Ministry of Public Security, Beijing 100038, China 6Beijing Botanical Garden, Beijing 100093, China Correspondence should be addressed to Ling Guo; [email protected] and Shiliang Zhou; [email protected] Received 21 September 2017; Revised 11 December 2017; Accepted 2 January 2018; Published 19 March 2018 Academic Editor: Fengjie Sun Copyright © 2018 Jiahui Sun et al. Tis is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Maleae consists of economically and ecologically important plants. However, there are considerable disputes on generic circumscription due to the lack of a reliable phylogeny at generic level. In this study, molecular phylogeny of 35 generally accepted genera in Maleae is established using 15 chloroplast regions. Gillenia isthemostbasalcladeofMaleae,followedbyKageneckia + Lindleya, Vauquelinia, and a typical radiation clade, the core Maleae, suggesting that the proposal of four subtribes is reasonable. -

Meadowsweet 2015
ONLINE SERIES MONOGRAPHS The Scientific Foundation for Herbal Medicinal Products Filipendulae ulmariae herba Meadowsweet 2015 www.escop.com The Scientific Foundation for Herbal Medicinal Products FILIPENDULAE ULMARIAE HERBA Black Cohosh 2015 ESCOP Monographs were first published in loose-leaf form progressively from 1996 to 1999 as Fascicules 1-6, each of 10 monographs © ESCOP 1996, 1997, 1999 Second Edition, completely revised and expanded © ESCOP 2003 Second Edition, Supplement 2009 © ESCOP 2009 ONLINE SERIES ISBN 978-1-901964-37-0 Filipendulae ulmariae herba - Meadowsweet © ESCOP 2015 Published by the European Scientific Cooperative on Phytotherapy (ESCOP) Notaries House, Chapel Street, Exeter EX1 1EZ, United Kingdom www.escop.com All rights reserved Except for the purposes of private study, research, criticism or review no part of this text may be reproduced, stored in a retrieval system or transmitted, in any form or by any means, without the written permission of the publisher. Important Note: Medical knowledge is ever-changing. As new research and clinical experience broaden our knowledge, changes in treatment may be required. In their efforts to provide information on the efficacy and safety of herbal drugs and herbal preparations, presented as a substantial overview together with summaries of relevant data, the authors of the material herein have consulted comprehensive sources believed to be reliable. However, in view of the possibility of human error by the authors or publisher of the work herein, or changes in medical knowledge, neither the authors nor the publisher, nor any other party involved in the preparation of this work, warrants that the information contained herein is in every respect accurate or complete, and they are not responsible for any errors or omissions or for results obtained by the use of such information. -
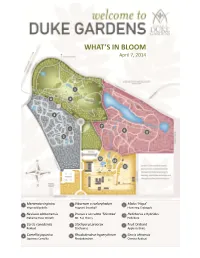
What's in Bloom
WHAT’S IN BLOOM April 7, 2014 5 4 6 2 7 1 9 8 3 12 10 11 1 Mertensia virginica 5 Viburnum x carlcephalum 9 Malus ‘Hopa’ Virginia Bluebells Fragrant Snowball Flowering Crabapple 2 Neviusia alabamensis 6 Prunus x serrulata ‘Shirotae’ 10 Helleborus x hybridus Alabama Snow Wreath Mt. Fuji Cherry Hellebore 3 Cercis canadensis 7 Stachyurus praecox 11 Fruit Orchard Redbud Stachyurus Apple cultivars 4 Camellia japonica 8 Rhododendron hyperythrum 12 Cercis chinensis Japanese Camellia Rhododendron Chinese Redbud WHAT’S IN BLOOM April 7, 2014 BLOMQUIST GARDEN OF NATIVE PLANTS Amelanchier arborea Common Serviceberry Sanguinaria canadensis Bloodroot Cornus florida Flowering Dogwood Stylophorum diphyllum Celandine Poppy Thalictrum thalictroides Rue Anemone Fothergilla major Fothergilla Trillium decipiens Chattahoochee River Trillium Hepatica nobilis Hepatica Trillium grandiflorum White Trillium Hexastylis virginica Wild Ginger Hexastylis minor Wild Ginger Trillium pusillum Dwarf Wakerobin Illicium floridanum Florida Anise Tree Trillium stamineum Blue Ridge Wakerobin Malus coronaria Sweet Crabapple Uvularia sessilifolia Sessileleaf Bellwort Mertensia virginica Virginia Bluebells Pachysandra procumbens Allegheny spurge Prunus americana American Plum DORIS DUKE CENTER GARDENS Camellia japonica Japanese Camellia Pulmonaria ‘Diana Clare’ Lungwort Cercis canadensis Redbud Prunus persica Flowering Peach Puschkinia scilloides Striped Squill Cercis chinensis Redbud Sanguinaria canadensis Bloodroot Clematis armandii Evergreen Clematis Spiraea prunifolia Bridalwreath -

Phylogenetic Inferences in Prunus (Rosaceae) Using Chloroplast Ndhf and Nuclear Ribosomal ITS Sequences 1Jun WEN* 2Scott T
Journal of Systematics and Evolution 46 (3): 322–332 (2008) doi: 10.3724/SP.J.1002.2008.08050 (formerly Acta Phytotaxonomica Sinica) http://www.plantsystematics.com Phylogenetic inferences in Prunus (Rosaceae) using chloroplast ndhF and nuclear ribosomal ITS sequences 1Jun WEN* 2Scott T. BERGGREN 3Chung-Hee LEE 4Stefanie ICKERT-BOND 5Ting-Shuang YI 6Ki-Oug YOO 7Lei XIE 8Joey SHAW 9Dan POTTER 1(Department of Botany, National Museum of Natural History, MRC 166, Smithsonian Institution, Washington, DC 20013-7012, USA) 2(Department of Biology, Colorado State University, Fort Collins, CO 80523, USA) 3(Korean National Arboretum, 51-7 Jikdongni Soheur-eup Pocheon-si Gyeonggi-do, 487-821, Korea) 4(UA Museum of the North and Department of Biology and Wildlife, University of Alaska Fairbanks, Fairbanks, AK 99775-6960, USA) 5(Key Laboratory of Plant Biodiversity and Biogeography, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650204, China) 6(Division of Life Sciences, Kangwon National University, Chuncheon 200-701, Korea) 7(State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China) 8(Department of Biological and Environmental Sciences, University of Tennessee, Chattanooga, TN 37403-2598, USA) 9(Department of Plant Sciences, MS 2, University of California, Davis, CA 95616, USA) Abstract Sequences of the chloroplast ndhF gene and the nuclear ribosomal ITS regions are employed to recon- struct the phylogeny of Prunus (Rosaceae), and evaluate the classification schemes of this genus. The two data sets are congruent in that the genera Prunus s.l. and Maddenia form a monophyletic group, with Maddenia nested within Prunus.