Systematic Summary for Genus and Subgenera by W
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Allium Albanicum (Amaryllidaceae), a New Species from Balkans and Its
A peer-reviewed open-access journal PhytoKeys 119: 117–136Allium (2019) albanicum (Amaryllidaceae), a new species from Balkans... 117 doi: 10.3897/phytokeys.119.30790 RESEARCH ARTICLE http://phytokeys.pensoft.net Launched to accelerate biodiversity research Allium albanicum (Amaryllidaceae), a new species from Balkans and its relationships with A. meteoricum Heldr. & Hausskn. ex Halácsy Salvatore Brullo1, Cristian Brullo2, Salvatore Cambria1, Giampietro Giusso del Galdo1, Cristina Salmeri2 1 Department of Biological, Geological and Environmental Sciences, Catania University, Via A. Longo 19, 95125 Catania, Italy 2 Department of Biological, Chemical and Pharmaceutical Sciences and Technologies (STEBICEF), Palermo University, Via Archirafi 38, 90123 Palermo, Italy Corresponding author: Cristina Salmeri ([email protected]) Academic editor: L. Peruzzi | Received 26 October 2018 | Accepted 9 January 2019 | Published 11 April 2019 Citation: Brullo S, Brullo C, Cambria S, Giusso del Galdo G, Salmeri C (2019) Allium albanicum (Amaryllidaceae), a new species from Balkans and its relationships with A. meteoricum Heldr. & Hausskn. ex Halácsy. PhytoKeys 119: 117–136. https://doi.org/10.3897/phytokeys.119.30790 Abstract A new species, Allium albanicum, is described and illustrated from Albania (Balkan Peninsula). It grows on serpentines or limestone in open rocky stands with a scattered distribution, mainly in mountain loca- tions. Previously, the populations of this geophyte were attributed to A. meteoricum Heldr. & Hausskn. ex Halácsy, described from a few localities of North and Central Greece. These two species indeed show close relationships, chiefly regarding some features of the spathe valves, inflorescence and floral parts. They also share the same diploid chromosome number 2n =16 and similar karyotype, while seed testa micro- sculptures and leaf anatomy reveal remarkable differences. -

AUTOMATIC FEEDBACK CONTROL in HUMAN BIOLOGY EEL 5934 Section 32513 Class Periods: MWF, Period 8, 3:00-3:50PM Location: Classroom Location Academic Term: Spring 2021
AUTOMATIC FEEDBACK CONTROL IN HUMAN BIOLOGY EEL 5934 Section 32513 Class Periods: MWF, period 8, 3:00-3:50PM Location: Classroom location Academic Term: Spring 2021 Instructor: Name: Jacob Hammer Email Address: [email protected] Office Phone Number: 3523924934 Office Hours: MWF hours available, office location Teaching Assistant/Peer Mentor/Supervised Teaching Student: Please contact through the Canvas website • Name, email address, office location, office hours • Name, email address, office location, office hours Course Description A course about the automatic feedback control principles that govern biological, biochemical, and genetic mechanisms underlying critical processes in human biology. The course concentrates on case studies, including the automatic feedback control principles that regulate vision, balance, heart rate, and various metabolic and immunologic processes in human biology. Course Pre-Requisites -Basic knowledge of control theory and linear algebra (EEL 4657C or EEL 4610 or equivalent) or instructor consent Course Objectives Introduce students to general principles of automatic control, as they are applied by natural pHenomena in human biology. Materials and Supply Fees List if applicable Required TextBooks and Software: • "Biomolecular Feedback Systems" • Domitilla Del VeccHio and RicHard M. Murray • Princeton University Press, Princeton, NJ, USA, 2014 • ISBN number (if course notes derived from various publisHed sources are used, provide information above for eacH source) (if course notes are developed by tHe instructor, -
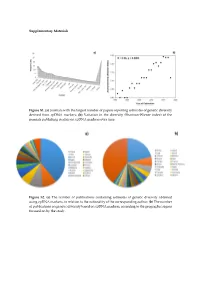
(A) Journals with the Largest Number of Papers Reporting Estimates Of
Supplementary Materials Figure S1. (a) Journals with the largest number of papers reporting estimates of genetic diversity derived from cpDNA markers; (b) Variation in the diversity (Shannon-Wiener index) of the journals publishing studies on cpDNA markers over time. Figure S2. (a) The number of publications containing estimates of genetic diversity obtained using cpDNA markers, in relation to the nationality of the corresponding author; (b) The number of publications on genetic diversity based on cpDNA markers, according to the geographic region focused on by the study. Figure S3. Classification of the angiosperm species investigated in the papers that analyzed genetic diversity using cpDNA markers: (a) Life mode; (b) Habitat specialization; (c) Geographic distribution; (d) Reproductive cycle; (e) Type of flower, and (f) Type of pollinator. Table S1. Plant species identified in the publications containing estimates of genetic diversity obtained from the use of cpDNA sequences as molecular markers. Group Family Species Algae Gigartinaceae Mazzaella laminarioides Angiospermae Typhaceae Typha laxmannii Angiospermae Typhaceae Typha orientalis Angiospermae Typhaceae Typha angustifolia Angiospermae Typhaceae Typha latifolia Angiospermae Araliaceae Eleutherococcus sessiliflowerus Angiospermae Polygonaceae Atraphaxis bracteata Angiospermae Plumbaginaceae Armeria pungens Angiospermae Aristolochiaceae Aristolochia kaempferi Angiospermae Polygonaceae Atraphaxis compacta Angiospermae Apocynaceae Lagochilus macrodontus Angiospermae Polygonaceae Atraphaxis -

H. Kiyosumiensis F
Hosta Species Update●The Hosta Library © W. George Schmid 2006 H. kiyosumiensis F. Maekawa 1935 Jornal of Japanese Botany 11:689; ic. f. 15 1935 キヨスミギボウシ = Kyosumi Giboshi H. kiyosumiensis var. petrophila F. Maekawa 1938 Divisiones et Plantae Novae Generis Hostae (2). J. Japanese Botany, 14:1:45–49. イワマ ギボウシ = Iwama Giboshi History and Nomenclature: In Japan this species is called Kyiosumi Giboshi, the “Kiyosumi (Mountain) Hosta.” The species epithet stems from the Latinized name of Kiyosumiyama (清澄山), a small mountain (380 m/about 1250 feet AMSL with a great view of Tokyo Bay) located on Boso Peninsula (Bōsō-hantō; 房総半島), which is located in south Chiba Prefecture (Chiba-ken; 千葉県) forming the eastern edge of Tokyo Bay. Maekawa established H. kiyosumiensis in 1935 and published further on the species in 1938, when he also described a new variety of the species as H. (kiyosumiensis var.) petrophila. This varietal epithet is derived from the Latin “liking (or) growing on rocks.” The Japanese name Iwama Giboshi (イワマ ギボウシ) has the same meaning. The latter is differentiated by minor morphological details caused by its different, rocky habitat in Yamashiro province (山城国; Yamashiro-no kuni), H. kiyosumiensis Maekawa 1935 (in situ) Otogawa (男川) River; Okazaki-shi (岡崎市; formerly Nukata-cho) Aichi Prefecture (愛知県; Aichi-ken) 1 an old province of Japan (today the southern part of Kyoto Prefecture). Maekawa (1938) gave a much abbreviated Latin description and the variety is here considered synonymous with the species. In fact, Maekawa (1969) no longer supported the varietal rank. N. Fujita (1976) confirmed H. kiyosumiensis as a species and added two morphs previously described by Maekawa (1940), i.e., H. -

Complete Chloroplast Genomes Shed Light on Phylogenetic
www.nature.com/scientificreports OPEN Complete chloroplast genomes shed light on phylogenetic relationships, divergence time, and biogeography of Allioideae (Amaryllidaceae) Ju Namgung1,4, Hoang Dang Khoa Do1,2,4, Changkyun Kim1, Hyeok Jae Choi3 & Joo‑Hwan Kim1* Allioideae includes economically important bulb crops such as garlic, onion, leeks, and some ornamental plants in Amaryllidaceae. Here, we reported the complete chloroplast genome (cpDNA) sequences of 17 species of Allioideae, fve of Amaryllidoideae, and one of Agapanthoideae. These cpDNA sequences represent 80 protein‑coding, 30 tRNA, and four rRNA genes, and range from 151,808 to 159,998 bp in length. Loss and pseudogenization of multiple genes (i.e., rps2, infA, and rpl22) appear to have occurred multiple times during the evolution of Alloideae. Additionally, eight mutation hotspots, including rps15-ycf1, rps16-trnQ-UUG, petG-trnW-CCA , psbA upstream, rpl32- trnL-UAG , ycf1, rpl22, matK, and ndhF, were identifed in the studied Allium species. Additionally, we present the frst phylogenomic analysis among the four tribes of Allioideae based on 74 cpDNA coding regions of 21 species of Allioideae, fve species of Amaryllidoideae, one species of Agapanthoideae, and fve species representing selected members of Asparagales. Our molecular phylogenomic results strongly support the monophyly of Allioideae, which is sister to Amaryllioideae. Within Allioideae, Tulbaghieae was sister to Gilliesieae‑Leucocoryneae whereas Allieae was sister to the clade of Tulbaghieae‑ Gilliesieae‑Leucocoryneae. Molecular dating analyses revealed the crown age of Allioideae in the Eocene (40.1 mya) followed by diferentiation of Allieae in the early Miocene (21.3 mya). The split of Gilliesieae from Leucocoryneae was estimated at 16.5 mya. -

Gondwanan Origin of Major Monocot Groups Inferred from Dispersal-Vicariance Analysis Kåre Bremer Uppsala University
Aliso: A Journal of Systematic and Evolutionary Botany Volume 22 | Issue 1 Article 3 2006 Gondwanan Origin of Major Monocot Groups Inferred from Dispersal-Vicariance Analysis Kåre Bremer Uppsala University Thomas Janssen Muséum National d'Histoire Naturelle Follow this and additional works at: http://scholarship.claremont.edu/aliso Part of the Botany Commons Recommended Citation Bremer, Kåre and Janssen, Thomas (2006) "Gondwanan Origin of Major Monocot Groups Inferred from Dispersal-Vicariance Analysis," Aliso: A Journal of Systematic and Evolutionary Botany: Vol. 22: Iss. 1, Article 3. Available at: http://scholarship.claremont.edu/aliso/vol22/iss1/3 Aliso 22, pp. 22-27 © 2006, Rancho Santa Ana Botanic Garden GONDWANAN ORIGIN OF MAJOR MONO COT GROUPS INFERRED FROM DISPERSAL-VICARIANCE ANALYSIS KARE BREMERl.3 AND THOMAS JANSSEN2 lDepartment of Systematic Botany, Evolutionary Biology Centre, Norbyvagen l8D, SE-752 36 Uppsala, Sweden; 2Museum National d'Histoire Naturelle, Departement de Systematique et Evolution, USM 0602: Taxonomie et collections, 16 rue Buffon, 75005 Paris, France 3Corresponding author ([email protected]) ABSTRACT Historical biogeography of major monocot groups was investigated by biogeographical analysis of a dated phylogeny including 79 of the 81 monocot families using the Angiosperm Phylogeny Group II (APG II) classification. Five major areas were used to describe the family distributions: Eurasia, North America, South America, Africa including Madagascar, and Australasia including New Guinea, New Caledonia, and New Zealand. In order to investigate the possible correspondence with continental breakup, the tree with its terminal distributions was fitted to the geological area cladogram «Eurasia, North America), (Africa, (South America, Australasia») and to alternative area cladograms using the TreeFitter program. -

Massonia Amoena (Asparagaceae, Scilloideae), a Striking New Species from the Eastern Cape, South Africa
Phytotaxa 181 (3): 121–137 ISSN 1179-3155 (print edition) www.mapress.com/phytotaxa/ PHYTOTAXA Copyright © 2014 Magnolia Press Article ISSN 1179-3163 (online edition) http://dx.doi.org/10.11646/phytotaxa.181.3.1 Massonia amoena (Asparagaceae, Scilloideae), a striking new species from the Eastern Cape, South Africa MARIO MARTÍNEZ-AZORÍN1,2, MICHAEL PINTER1, GERFRIED DEUTSCH1, ANDREAS BRUDERMANN1, ANTHONY P. DOLD3, MANUEL B. CRESPO2, MARTIN PFOSSER4 & WOLFGANG WETSCHNIG1* 1Institute of Plant Sciences, NAWI Graz, Karl-Franzens-University Graz, Holteigasse 6, A-8010 Graz, Austria; e-mail: wolfgang.wet- [email protected] 2CIBIO (Instituto Universitario de la Biodiversidad), Universidad de Alicante, P. O. Box 99, E-03080 Alicante, Spain. 3Selmar Schonland Herbarium, Department of Botany, Rhodes University, Grahamstown 6140 South Africa. 4Biocenter Linz, J.-W.-Klein-Str. 73, A-4040 Linz, Austria. *author for correspondence Abstract As part of an ongoing study towards a taxonomic revision of the genus Massonia Houtt., a new species, Massonia amoena Mart.-Azorín, M.Pinter & Wetschnig, is here described from the Eastern Cape Province of South Africa. This new species is characterized by the leaves bearing heterogeneous circular to elongate pustules and the strongly reflexed perigone seg- ments at anthesis. It is at first sight related to Massonia jasminiflora Burch. ex Baker, M. wittebergensis U.Müll.-Doblies & D.Müll.-Doblies and M. saniensis Wetschnig, Mart.-Azorín & M.Pinter, but differs in vegetative and floral characters, as well as in its allopatric distribution. A complete morphological description of the new species and data on biology, habitat, and distribution are presented. Key words: flora, Hyacinthaceae, Massonieae, Southern Africa, taxonomy Introduction Hyacinthaceae sensu APG (2003) includes ca. -

Full of Beans: a Study on the Alignment of Two Flowering Plants Classification Systems
Full of beans: a study on the alignment of two flowering plants classification systems Yi-Yun Cheng and Bertram Ludäscher School of Information Sciences, University of Illinois at Urbana-Champaign, USA {yiyunyc2,ludaesch}@illinois.edu Abstract. Advancements in technologies such as DNA analysis have given rise to new ways in organizing organisms in biodiversity classification systems. In this paper, we examine the feasibility of aligning two classification systems for flowering plants using a logic-based, Region Connection Calculus (RCC-5) ap- proach. The older “Cronquist system” (1981) classifies plants using their mor- phological features, while the more recent Angiosperm Phylogeny Group IV (APG IV) (2016) system classifies based on many new methods including ge- nome-level analysis. In our approach, we align pairwise concepts X and Y from two taxonomies using five basic set relations: congruence (X=Y), inclusion (X>Y), inverse inclusion (X<Y), overlap (X><Y), and disjointness (X!Y). With some of the RCC-5 relationships among the Fabaceae family (beans family) and the Sapindaceae family (maple family) uncertain, we anticipate that the merging of the two classification systems will lead to numerous merged solutions, so- called possible worlds. Our research demonstrates how logic-based alignment with ambiguities can lead to multiple merged solutions, which would not have been feasible when aligning taxonomies, classifications, or other knowledge or- ganization systems (KOS) manually. We believe that this work can introduce a novel approach for aligning KOS, where merged possible worlds can serve as a minimum viable product for engaging domain experts in the loop. Keywords: taxonomy alignment, KOS alignment, interoperability 1 Introduction With the advent of large-scale technologies and datasets, it has become increasingly difficult to organize information using a stable unitary classification scheme over time. -

Generic Classification of Amaryllidaceae Tribe Hippeastreae Nicolás García,1 Alan W
TAXON 2019 García & al. • Genera of Hippeastreae SYSTEMATICS AND PHYLOGENY Generic classification of Amaryllidaceae tribe Hippeastreae Nicolás García,1 Alan W. Meerow,2 Silvia Arroyo-Leuenberger,3 Renata S. Oliveira,4 Julie H. Dutilh,4 Pamela S. Soltis5 & Walter S. Judd5 1 Herbario EIF & Laboratorio de Sistemática y Evolución de Plantas, Facultad de Ciencias Forestales y de la Conservación de la Naturaleza, Universidad de Chile, Av. Santa Rosa 11315, La Pintana, Santiago, Chile 2 USDA-ARS-SHRS, National Germplasm Repository, 13601 Old Cutler Rd., Miami, Florida 33158, U.S.A. 3 Instituto de Botánica Darwinion, Labardén 200, CC 22, B1642HYD, San Isidro, Buenos Aires, Argentina 4 Departamento de Biologia Vegetal, Instituto de Biologia, Universidade Estadual de Campinas, Postal Code 6109, 13083-970 Campinas, SP, Brazil 5 Florida Museum of Natural History, University of Florida, Gainesville, Florida 32611, U.S.A. Address for correspondence: Nicolás García, [email protected] DOI https://doi.org/10.1002/tax.12062 Abstract A robust generic classification for Amaryllidaceae has remained elusive mainly due to the lack of unequivocal diagnostic characters, a consequence of highly canalized variation and a deeply reticulated evolutionary history. A consensus classification is pro- posed here, based on recent molecular phylogenetic studies, morphological and cytogenetic variation, and accounting for secondary criteria of classification, such as nomenclatural stability. Using the latest sutribal classification of Hippeastreae (Hippeastrinae and Traubiinae) as a foundation, we propose the recognition of six genera, namely Eremolirion gen. nov., Hippeastrum, Phycella s.l., Rhodolirium s.str., Traubia, and Zephyranthes s.l. A subgeneric classification is suggested for Hippeastrum and Zephyranthes to denote putative subclades. -

Reducing Deer Damage in Landscapes Part 2
Reducing Deer Damage in the Landscape D Coetzee Public Domain bestfriendthemom Dusty Hancock CC BY-NC-ND 2.0 Chatham Master Gardener Volunteer Matt Jones Horticulture Agent NC Cooperative Extension - Chatham County Center Plant Selection Deer Candy • Aucuba • Hosta • Arborvitae • Indian Hawthorn • Azalea • Ivy • Blueberry • Chionanthus, • Clematis Malus, Prunus, • Daylily Pyrus • Euonymus • Redbuds • Fatsia • Roses Muhenbergia capillaris Pink Muhly Grass (Poaceae) Andrea Laine Jim Robbins CC BY NC 4.0 CC BY-NC-ND 4.0 Muhlenbergia capillaris Pink Muhly Grass (Poaceae) Full Sun Moist to very dry 1-3’ x 1-3’ Fall Susan Strine Fall-Winter CC BY 2.0 Schizachyrium scoparium Little Blue Stem (Poaceae) Jim Robbins Joshua Mayer Jim Robbins Montreais CC BY-SA 2.0 DE CC BY-NC-ND 4.0 CC BY-NC-ND 4.0 CC BY-SA 3.0 Schizachyrium scoparium Little Blue Stem (Poaceae) Full Sun Moist to dry Good drainage 1-4’ x 18”-2’ Summer-Fall Susan Strine Summer-Fall Jim RobbinsCC BY 2.0 CC BY-NC-ND 4.0 Chasmanthium latifolium River Oats (Poaceae) Klasse im Garten Anne McCormack CC BY 2.0 CC BY-NC 2.0 Chasmanthium latifolium River Oats (Poaceae) Part shade to dappled sun Moist Occasionally wet 2-5’ x 1-2’ Summer-Fall Anne McCormack Summer-Fall CC BY-NC 2.0 Myrica cerifera (Myricaceae) Common Wax Myrtle Forest and Kim Starr CC BY 2.0 Forest and Kim Starr CC BY 2.0 Myrica cerifera (Myricaceae) Common Wax Myrtle Jim Robbins CC BY-NC-ND 4.0 Jim Robbins Jim Robbins CC BY-NC-ND 4.0 CC BY-NC-ND 4.0 Great for urban soils, full sun to part shade. -

Mini Data Sheet on Asparagus Asparagoides (Asparagaceae)
EPPO, 2013 Mini data sheet on Asparagus asparagoides (Asparagaceae) Added in 2012 – Deleted in 2013 Reasons for deletion: Asparagus asparagoides was added to the EPPO Alert List in 2012 but as no immediate risk was perceived, it was transferred to the Observation List in 2013. Why Asparagus asparagoides (Asparagaceae) is a rhizomatous perennial climbing vine originating from South Africa. One of its English common names is “bridal creeper”. This species is invasive in Australia. It is used as an ornamental plant in the EPPO region, and is listed as an invasive alien plant in Spain, but is also present in other EPPO member countries. Considering the invasive behavior of this species elsewhere in the world as well as in EPPO countries, it is considered that Mediterranean and Macaronesian countries may be at risk, and that the species should usefully be monitored. Geographical distribution EPPO region: France (including Corse), Italy (Sicilia), Malta, Morocco, Portugal (Azores, Madeira), Spain (including Islas Canarias), Tunisia. Note: The species had erroneously been indicated as present in Slovenia (from an incorrect interpretation of Jogan, 2005). Africa (native): Ethiopia, Kenya, Lesotho, Malawi, Morocco, Namibia, South Africa, Swaziland, Tanzania, Tunisia, Uganda, Zimbabwe. North America: Mexico, USA (California, Hawaii (East Maui)). South and Central America: Argentina, Guatemala, Uruguay. Oceania (invasive): Australia (New South Wales, Queensland, South Australia, Tasmania, Victoria, Western Australia), New Zealand. Morphology A. asparagoides is a geophyte with a perennial cylindrical, slender (about 5 mm wide), branching rhizome, growing parallel to the soil surface, bearing fleshy tubers (25–42 mm long and 8–20 mm wide). It produces thin shoots, slightly woody at the base and up to 6 m long when support is available. -

(Monocot): the Story of a Gene Family Expansion Alberto Cenci, Nathalie Chantret, Mathieu Rouard
Glycosyltransferase Family 61 in Liliopsida (Monocot): The Story of a Gene Family Expansion Alberto Cenci, Nathalie Chantret, Mathieu Rouard To cite this version: Alberto Cenci, Nathalie Chantret, Mathieu Rouard. Glycosyltransferase Family 61 in Liliopsida (Monocot): The Story of a Gene Family Expansion. Frontiers in Plant Science, Frontiers, 2018, 9, 10.3389/fpls.2018.01843. hal-02621636 HAL Id: hal-02621636 https://hal.inrae.fr/hal-02621636 Submitted on 26 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License fpls-09-01843 December 8, 2018 Time: 15:6 # 1 ORIGINAL RESEARCH published: 11 December 2018 doi: 10.3389/fpls.2018.01843 Glycosyltransferase Family 61 in Liliopsida (Monocot): The Story of a Gene Family Expansion Alberto Cenci1*, Nathalie Chantret2 and Mathieu Rouard1 1 Bioversity International, Parc Scientifique Agropolis II, Montpellier, France, 2 AGAP, INRA, CIRAD, Université de Montpellier, Montpellier, France Plant cell walls play a fundamental role in several plant traits and also influence crop use as livestock nutrition or biofuel production. The Glycosyltransferase family 61 (GT61) is involved in the synthesis of cell wall xylans.