Jan Dvorak (As Quickly Written Down by a Person with Poor Hearing…Me)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Florística E Fitossociologia Da Restinga Herbáceo-Arbustiva Do Morro Dos Conventos, Araranguá, Sc
UNIVERSIDADE DO EXTREMO SUL CATARINENSE – UNESC PROGRAMA DE PÓS-GRADUAÇÃO EM CIÊNCIAS AMBIENTAIS – PPGCA (MESTRADO) FLORÍSTICA E FITOSSOCIOLOGIA DA RESTINGA HERBÁCEO-ARBUSTIVA DO MORRO DOS CONVENTOS, ARARANGUÁ, SC Rosabel Bertolin Daniel CRICIÚMA (SC), 2006 ii FLORÍSTICA E FITOSSOCIOLOGIA DA RESTINGA HERBÁCEO-ARBUSTIVA DO MORRO DOS CONVENTOS, ARARANGUÁ, SC Rosabel Bertolin Daniel Dissertação apresentada ao Programa de Pós-Graduação em Ciências Ambientais – PPGCA, da Universidade do Extremo Sul Catarinense – UNESC, como requisito parcial para a obtenção do titulo de Mestre em Ciências Ambientais. Área de Concentração: Ecologia, manejo e gestão de ambientes naturais ou impactados Orientador: Prof. Dr. Jairo José Zocche CRICIÚMA (SC), 2006 iii Dados Internacionais de Catalogação na Publicação D184f Daniel, Rosabel Bertolin. Florística e fitossociologia da restinga herbáceo-arbustiva do Morro dos Conventos, Araranguá,SC/ Rosabel Bertolin Daniel ; orientador : Jairo José Zocche . – Criciúma : Ed. do autor, 2006. 74 f. : il. ; 30 cm. Dissertação (Mestrado) - Universidade do Extremo Sul Catarinense. Programa de Pós-Graduação em Ciências Ambientais, Criciúma, 2006. 1. Restingas - Vegetação. 2. Vegetação – Morro dos Conventos, SC. I. Título. CDD. 21ª ed. 581.751098164 Bibliotecária: Flávia Cardoso – CRB 14/840 Biblioteca Central Prof. Eurico Back – UNESC iv “Qualquer caminho é apenas um caminho e não constitui insulto algum – para si mesmo ou para os outros – abandoná-lo quando assim ordena seu coração. (...) Olhe cada caminho com cuidado e atenção. Tente-o tantas vezes quantas julgar necessárias... Então, faça a si mesmo e apenas a si mesmo uma pergunta: possui esse caminho um coração? Em caso afirmativo, o caminho é bom. Caso contrário, esse caminho não possui importância alguma”. -

Natural Heritage Program List of Rare Plant Species of North Carolina 2016
Natural Heritage Program List of Rare Plant Species of North Carolina 2016 Revised February 24, 2017 Compiled by Laura Gadd Robinson, Botanist John T. Finnegan, Information Systems Manager North Carolina Natural Heritage Program N.C. Department of Natural and Cultural Resources Raleigh, NC 27699-1651 www.ncnhp.org C ur Alleghany rit Ashe Northampton Gates C uc Surry am k Stokes P d Rockingham Caswell Person Vance Warren a e P s n Hertford e qu Chowan r Granville q ot ui a Mountains Watauga Halifax m nk an Wilkes Yadkin s Mitchell Avery Forsyth Orange Guilford Franklin Bertie Alamance Durham Nash Yancey Alexander Madison Caldwell Davie Edgecombe Washington Tyrrell Iredell Martin Dare Burke Davidson Wake McDowell Randolph Chatham Wilson Buncombe Catawba Rowan Beaufort Haywood Pitt Swain Hyde Lee Lincoln Greene Rutherford Johnston Graham Henderson Jackson Cabarrus Montgomery Harnett Cleveland Wayne Polk Gaston Stanly Cherokee Macon Transylvania Lenoir Mecklenburg Moore Clay Pamlico Hoke Union d Cumberland Jones Anson on Sampson hm Duplin ic Craven Piedmont R nd tla Onslow Carteret co S Robeson Bladen Pender Sandhills Columbus New Hanover Tidewater Coastal Plain Brunswick THE COUNTIES AND PHYSIOGRAPHIC PROVINCES OF NORTH CAROLINA Natural Heritage Program List of Rare Plant Species of North Carolina 2016 Compiled by Laura Gadd Robinson, Botanist John T. Finnegan, Information Systems Manager North Carolina Natural Heritage Program N.C. Department of Natural and Cultural Resources Raleigh, NC 27699-1651 www.ncnhp.org This list is dynamic and is revised frequently as new data become available. New species are added to the list, and others are dropped from the list as appropriate. -
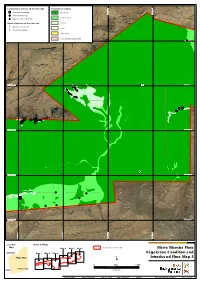
Additional Information
Current Survey Introduced Flora Records Vegetation Condition *Acetosa vesicaria Excellent 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 -

Wiregrass (Aristida Stricta)
Wiregrass (Aristida stricta) For definitions of botanical terms, visit en.wikipedia.org/wiki/Glossary_of_botanical_terms. Wiregrass is a perennial bunchgrass found in scrub, pinelands and coastal uplands throughout much of Florida. It is the dominant groundcover species in longleaf pine savannas and is a primary food source for gopher tortoises. Birds and small wildlife eat the seeds. Historically, cattle grazed on Wiregrass’s tender new growth. Wiregrass flowers are tiny and brown. They are born on spikelike terminal panicles Flower stalks are elongated and extend above the leaves. Leaf blades are long, thin and rolled inward, giving them a wiry appearance (hence the common name). They are erect and green when young Photo courtesy of Alan Cressler, Lady Bird Johnson Wildflower Center and begin to arch and turn brown as they age. Its fruits are small, yellowish caryopses. Seeds may be dispersed by wind, gravity or on the fur of passing animals. The genus name Aristida is from the Latin arista, meaning “awn” and referring to the three awns or bristle-like structures that extend from the florets. (An alternative common name is Pineland threeawn.) The species epithet, stricta, is from the Latin strictus, meaning straight or erect. Family: Poaceae (Grass family) Native range: Nearly throughout To see where natural populations of Wiregrass have been vouchered, visit www.florida.plantatlas.usf.edu. Hardiness: Zones 8A–10B Lifespan: Perennial Soil: Moist to dry, well-drained sandy soils Exposure: Full sun to partial shade Growth habit: 1–3’+ tall and equally wide Propagation: Division, seed Garden tips: Wiregrass is fast-growing and tolerant of drought conditions and low-nutrient soils. -
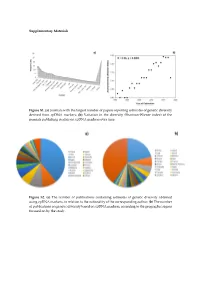
(A) Journals with the Largest Number of Papers Reporting Estimates Of
Supplementary Materials Figure S1. (a) Journals with the largest number of papers reporting estimates of genetic diversity derived from cpDNA markers; (b) Variation in the diversity (Shannon-Wiener index) of the journals publishing studies on cpDNA markers over time. Figure S2. (a) The number of publications containing estimates of genetic diversity obtained using cpDNA markers, in relation to the nationality of the corresponding author; (b) The number of publications on genetic diversity based on cpDNA markers, according to the geographic region focused on by the study. Figure S3. Classification of the angiosperm species investigated in the papers that analyzed genetic diversity using cpDNA markers: (a) Life mode; (b) Habitat specialization; (c) Geographic distribution; (d) Reproductive cycle; (e) Type of flower, and (f) Type of pollinator. Table S1. Plant species identified in the publications containing estimates of genetic diversity obtained from the use of cpDNA sequences as molecular markers. Group Family Species Algae Gigartinaceae Mazzaella laminarioides Angiospermae Typhaceae Typha laxmannii Angiospermae Typhaceae Typha orientalis Angiospermae Typhaceae Typha angustifolia Angiospermae Typhaceae Typha latifolia Angiospermae Araliaceae Eleutherococcus sessiliflowerus Angiospermae Polygonaceae Atraphaxis bracteata Angiospermae Plumbaginaceae Armeria pungens Angiospermae Aristolochiaceae Aristolochia kaempferi Angiospermae Polygonaceae Atraphaxis compacta Angiospermae Apocynaceae Lagochilus macrodontus Angiospermae Polygonaceae Atraphaxis -

H. Kiyosumiensis F
Hosta Species Update●The Hosta Library © W. George Schmid 2006 H. kiyosumiensis F. Maekawa 1935 Jornal of Japanese Botany 11:689; ic. f. 15 1935 キヨスミギボウシ = Kyosumi Giboshi H. kiyosumiensis var. petrophila F. Maekawa 1938 Divisiones et Plantae Novae Generis Hostae (2). J. Japanese Botany, 14:1:45–49. イワマ ギボウシ = Iwama Giboshi History and Nomenclature: In Japan this species is called Kyiosumi Giboshi, the “Kiyosumi (Mountain) Hosta.” The species epithet stems from the Latinized name of Kiyosumiyama (清澄山), a small mountain (380 m/about 1250 feet AMSL with a great view of Tokyo Bay) located on Boso Peninsula (Bōsō-hantō; 房総半島), which is located in south Chiba Prefecture (Chiba-ken; 千葉県) forming the eastern edge of Tokyo Bay. Maekawa established H. kiyosumiensis in 1935 and published further on the species in 1938, when he also described a new variety of the species as H. (kiyosumiensis var.) petrophila. This varietal epithet is derived from the Latin “liking (or) growing on rocks.” The Japanese name Iwama Giboshi (イワマ ギボウシ) has the same meaning. The latter is differentiated by minor morphological details caused by its different, rocky habitat in Yamashiro province (山城国; Yamashiro-no kuni), H. kiyosumiensis Maekawa 1935 (in situ) Otogawa (男川) River; Okazaki-shi (岡崎市; formerly Nukata-cho) Aichi Prefecture (愛知県; Aichi-ken) 1 an old province of Japan (today the southern part of Kyoto Prefecture). Maekawa (1938) gave a much abbreviated Latin description and the variety is here considered synonymous with the species. In fact, Maekawa (1969) no longer supported the varietal rank. N. Fujita (1976) confirmed H. kiyosumiensis as a species and added two morphs previously described by Maekawa (1940), i.e., H. -

Department of the Interior Fish and Wildlife Service
Tuesday, February 10, 2009 Part II Department of the Interior Fish and Wildlife Service 50 CFR Part 17 Endangered and Threatened Wildlife and Plants; Determination of Endangered Status for Reticulated Flatwoods Salamander; Designation of Critical Habitat for Frosted Flatwoods Salamander and Reticulated Flatwoods Salamander; Final Rule VerDate Nov<24>2008 14:17 Feb 09, 2009 Jkt 217001 PO 00000 Frm 00001 Fmt 4717 Sfmt 4717 E:\FR\FM\10FER2.SGM 10FER2 erowe on PROD1PC63 with RULES_2 6700 Federal Register / Vol. 74, No. 26 / Tuesday, February 10, 2009 / Rules and Regulations DEPARTMENT OF THE INTERIOR during normal business hours, at U.S. Register on or before July 30, 2008, with Fish and Wildlife Service, Mississippi the final critical habitat rule to be Fish and Wildlife Service Fish and Wildlife Office, 6578 Dogwood submitted for publication in the Federal View Parkway, Jackson, MS 39213. Register by January 30, 2009. The 50 CFR Part 17 FOR FURTHER INFORMATION CONTACT: Ray revised proposed rule was signed on [FWS–R4–ES–2008–0082; MO 9921050083– Aycock, Field Supervisor, U.S. Fish and and delivered to the Federal Register on B2] Wildlife Service, Mississippi Field July 30, 2008, and it subsequently Office, 6578 Dogwood View Parkway, published on August 13, 2008 (73 FR RIN 1018–AU85 Jackson, MS 39213; telephone: 601– 47258). We also published supplemental information on the Endangered and Threatened Wildlife 321–1122; facsimile: 601–965–4340. If proposed rule to maintain the status of and Plants; Determination of you use a telecommunications device the frosted flatwoods salamander as Endangered Status for Reticulated for the deaf (TDD), call the Federal Information Relay Service (FIRS) at threatened (73 FR 54125; September 18, Flatwoods Salamander; Designation of 2008). -

GENOME EVOLUTION in MONOCOTS a Dissertation
GENOME EVOLUTION IN MONOCOTS A Dissertation Presented to The Faculty of the Graduate School At the University of Missouri In Partial Fulfillment Of the Requirements for the Degree Doctor of Philosophy By Kate L. Hertweck Dr. J. Chris Pires, Dissertation Advisor JULY 2011 The undersigned, appointed by the dean of the Graduate School, have examined the dissertation entitled GENOME EVOLUTION IN MONOCOTS Presented by Kate L. Hertweck A candidate for the degree of Doctor of Philosophy And hereby certify that, in their opinion, it is worthy of acceptance. Dr. J. Chris Pires Dr. Lori Eggert Dr. Candace Galen Dr. Rose‐Marie Muzika ACKNOWLEDGEMENTS I am indebted to many people for their assistance during the course of my graduate education. I would not have derived such a keen understanding of the learning process without the tutelage of Dr. Sandi Abell. Members of the Pires lab provided prolific support in improving lab techniques, computational analysis, greenhouse maintenance, and writing support. Team Monocot, including Dr. Mike Kinney, Dr. Roxi Steele, and Erica Wheeler were particularly helpful, but other lab members working on Brassicaceae (Dr. Zhiyong Xiong, Dr. Maqsood Rehman, Pat Edger, Tatiana Arias, Dustin Mayfield) all provided vital support as well. I am also grateful for the support of a high school student, Cady Anderson, and an undergraduate, Tori Docktor, for their assistance in laboratory procedures. Many people, scientist and otherwise, helped with field collections: Dr. Travis Columbus, Hester Bell, Doug and Judy McGoon, Julie Ketner, Katy Klymus, and William Alexander. Many thanks to Barb Sonderman for taking care of my greenhouse collection of many odd plants brought back from the field. -

ISB: Atlas of Florida Vascular Plants
Longleaf Pine Preserve Plant List Acanthaceae Asteraceae Wild Petunia Ruellia caroliniensis White Aster Aster sp. Saltbush Baccharis halimifolia Adoxaceae Begger-ticks Bidens mitis Walter's Viburnum Viburnum obovatum Deer Tongue Carphephorus paniculatus Pineland Daisy Chaptalia tomentosa Alismataceae Goldenaster Chrysopsis gossypina Duck Potato Sagittaria latifolia Cow Thistle Cirsium horridulum Tickseed Coreopsis leavenworthii Altingiaceae Elephant's foot Elephantopus elatus Sweetgum Liquidambar styraciflua Oakleaf Fleabane Erigeron foliosus var. foliosus Fleabane Erigeron sp. Amaryllidaceae Prairie Fleabane Erigeron strigosus Simpson's rain lily Zephyranthes simpsonii Fleabane Erigeron vernus Dog Fennel Eupatorium capillifolium Anacardiaceae Dog Fennel Eupatorium compositifolium Winged Sumac Rhus copallinum Dog Fennel Eupatorium spp. Poison Ivy Toxicodendron radicans Slender Flattop Goldenrod Euthamia caroliniana Flat-topped goldenrod Euthamia minor Annonaceae Cudweed Gamochaeta antillana Flag Pawpaw Asimina obovata Sneezeweed Helenium pinnatifidum Dwarf Pawpaw Asimina pygmea Blazing Star Liatris sp. Pawpaw Asimina reticulata Roserush Lygodesmia aphylla Rugel's pawpaw Deeringothamnus rugelii Hempweed Mikania cordifolia White Topped Aster Oclemena reticulata Apiaceae Goldenaster Pityopsis graminifolia Button Rattlesnake Master Eryngium yuccifolium Rosy Camphorweed Pluchea rosea Dollarweed Hydrocotyle sp. Pluchea Pluchea spp. Mock Bishopweed Ptilimnium capillaceum Rabbit Tobacco Pseudognaphalium obtusifolium Blackroot Pterocaulon virgatum -

Reducing Deer Damage in Landscapes Part 2
Reducing Deer Damage in the Landscape D Coetzee Public Domain bestfriendthemom Dusty Hancock CC BY-NC-ND 2.0 Chatham Master Gardener Volunteer Matt Jones Horticulture Agent NC Cooperative Extension - Chatham County Center Plant Selection Deer Candy • Aucuba • Hosta • Arborvitae • Indian Hawthorn • Azalea • Ivy • Blueberry • Chionanthus, • Clematis Malus, Prunus, • Daylily Pyrus • Euonymus • Redbuds • Fatsia • Roses Muhenbergia capillaris Pink Muhly Grass (Poaceae) Andrea Laine Jim Robbins CC BY NC 4.0 CC BY-NC-ND 4.0 Muhlenbergia capillaris Pink Muhly Grass (Poaceae) Full Sun Moist to very dry 1-3’ x 1-3’ Fall Susan Strine Fall-Winter CC BY 2.0 Schizachyrium scoparium Little Blue Stem (Poaceae) Jim Robbins Joshua Mayer Jim Robbins Montreais CC BY-SA 2.0 DE CC BY-NC-ND 4.0 CC BY-NC-ND 4.0 CC BY-SA 3.0 Schizachyrium scoparium Little Blue Stem (Poaceae) Full Sun Moist to dry Good drainage 1-4’ x 18”-2’ Summer-Fall Susan Strine Summer-Fall Jim RobbinsCC BY 2.0 CC BY-NC-ND 4.0 Chasmanthium latifolium River Oats (Poaceae) Klasse im Garten Anne McCormack CC BY 2.0 CC BY-NC 2.0 Chasmanthium latifolium River Oats (Poaceae) Part shade to dappled sun Moist Occasionally wet 2-5’ x 1-2’ Summer-Fall Anne McCormack Summer-Fall CC BY-NC 2.0 Myrica cerifera (Myricaceae) Common Wax Myrtle Forest and Kim Starr CC BY 2.0 Forest and Kim Starr CC BY 2.0 Myrica cerifera (Myricaceae) Common Wax Myrtle Jim Robbins CC BY-NC-ND 4.0 Jim Robbins Jim Robbins CC BY-NC-ND 4.0 CC BY-NC-ND 4.0 Great for urban soils, full sun to part shade. -

Vicariance, Climate Change, Anatomy and Phylogeny of Restionaceae
Botanical Journal of the Linnean Society (2000), 134: 159–177. With 12 figures doi:10.1006/bojl.2000.0368, available online at http://www.idealibrary.com on Under the microscope: plant anatomy and systematics. Edited by P. J. Rudall and P. Gasson Vicariance, climate change, anatomy and phylogeny of Restionaceae H. P. LINDER FLS Bolus Herbarium, University of Cape Town, Rondebosch 7701, South Africa Cutler suggested almost 30 years ago that there was convergent evolution between African and Australian Restionaceae in the distinctive culm anatomical features of Restionaceae. This was based on his interpretation of the homologies of the anatomical features, and these are here tested against a ‘supertree’ phylogeny, based on three separate phylogenies. The first is based on morphology and includes all genera; the other two are based on molecular sequences from the chloroplast genome; one covers the African genera, and the other the Australian genera. This analysis corroborates Cutler’s interpretation of convergent evolution between African and Australian Restionaceae. However, it indicates that for the Australian genera, the evolutionary pathway of the culm anatomy is much more complex than originally thought. In the most likely scenario, the ancestral Restionaceae have protective cells derived from the chlorenchyma. These persist in African Restionaceae, but are soon lost in Australian Restionaceae. Pillar cells and sclerenchyma ribs evolve early in the diversification of Australian Restionaceae, but are secondarily lost numerous times. In some of the reduction cases, the result is a very simple culm anatomy, which Cutler had interpreted as a primitively simple culm type, while in other cases it appears as if the functions of the ribs and pillars may have been taken over by a new structure, protective cells developed from epidermal, rather than chlorenchyma, cells. -

ATLAS of FLORIDA PLANTS - 7/29/19 Lake County Native Species
ATLAS OF FLORIDA PLANTS - 7/29/19 Lake County Native Species Scientific_Name Common_Name Endemic State US 1 Abutilon hulseanum MAUVE N 2 Acalypha gracilens SLENDER THREESEED MERCURY N 3 Acalypha ostryifolia PINELAND THREESEED MERCURY N 4 Acer negundo BOXELDER N 5 Acer rubrum RED MAPLE N 6 Acrolejeunea heterophylla 7 Acrostichum danaeifolium GIANT LEATHER FERN N 8 Aeschynomene americana SHYLEAF N 9 Aeschynomene viscidula STICKY JOINTVETCH N 10 Aesculus pavia RED BUCKEYE N 11 Agalinis fasciculata BEACH FALSE FOXGLOVE N 12 Agalinis filifolia SEMINOLE FALSE FOXGLOVE N 13 Agalinis linifolia FLAXLEAF FALSE FOXGLOVE N 14 Agalinis plukenetii PLUKENET'S FALSE FOXGLOVE N 15 Agarista populifolia FLORIDA HOBBLEBUSH; PIPESTEM N 16 Ageratina jucunda HAMMOCK SNAKEROOT N 17 Aletris lutea YELLOW COLICROOT N 18 Allium canadense var. canadense MEADOW GARLIC N 19 Amaranthus australis SOUTHERN AMARANTH N 20 Amblystegium serpens 21 Ambrosia artemisiifolia COMMON RAGWEED N 22 Amorpha fruticosa BASTARD FALSE INDIGO N 23 Amorpha herbacea var. herbacea CLUSTERSPIKE FALSE INDIGO N 24 Amphicarpum muehlenbergianum BLUE MAIDENCANE N 25 Amsonia ciliata FRINGED BLUESTAR N 26 Andropogon brachystachyus SHORTSPIKE BLUESTEM N 27 Andropogon floridanus FLORIDA BLUESTEM N 28 Andropogon glomeratus var. glaucopsis PURPLE BLUESTEM N 29 Andropogon glomeratus var. hirsutior BUSHY BLUESTEM N 30 Andropogon glomeratus var. pumilus BUSHY BLUESTEM N 31 Andropogon gyrans ELLIOTT'S BLUESTEM N 32 Andropogon longiberbis HAIRY BLUESTEM N 33 Andropogon ternarius SPLITBEARD BLUESTEM N 34 Andropogon