Widespread Soil Bacterium That Oxidizes Atmospheric Methane
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Folic Acid Antagonists: Antimicrobial and Immunomodulating Mechanisms and Applications
International Journal of Molecular Sciences Review Folic Acid Antagonists: Antimicrobial and Immunomodulating Mechanisms and Applications Daniel Fernández-Villa 1, Maria Rosa Aguilar 1,2 and Luis Rojo 1,2,* 1 Instituto de Ciencia y Tecnología de Polímeros, Consejo Superior de Investigaciones Científicas, CSIC, 28006 Madrid, Spain; [email protected] (D.F.-V.); [email protected] (M.R.A.) 2 Consorcio Centro de Investigación Biomédica en Red de Bioingeniería, Biomateriales y Nanomedicina, 28029 Madrid, Spain * Correspondence: [email protected]; Tel.: +34-915-622-900 Received: 18 September 2019; Accepted: 7 October 2019; Published: 9 October 2019 Abstract: Bacterial, protozoan and other microbial infections share an accelerated metabolic rate. In order to ensure a proper functioning of cell replication and proteins and nucleic acids synthesis processes, folate metabolism rate is also increased in these cases. For this reason, folic acid antagonists have been used since their discovery to treat different kinds of microbial infections, taking advantage of this metabolic difference when compared with human cells. However, resistances to these compounds have emerged since then and only combined therapies are currently used in clinic. In addition, some of these compounds have been found to have an immunomodulatory behavior that allows clinicians using them as anti-inflammatory or immunosuppressive drugs. Therefore, the aim of this review is to provide an updated state-of-the-art on the use of antifolates as antibacterial and immunomodulating agents in the clinical setting, as well as to present their action mechanisms and currently investigated biomedical applications. Keywords: folic acid antagonists; antifolates; antibiotics; antibacterials; immunomodulation; sulfonamides; antimalarial 1. -

One Carbon Metabolism and Its Clinical Significance
One carbon metabolism and its clinical significance Dr. Kiran Meena Department of Biochemistry Class 5 : 10-10-2019 (8:00 to 9:00 AM ) Specific Learning Objectives Describe roles of folic acid, cobalamin and S-adenosylmethionine (SAM) in transfer of one carbon units between molecules, and apply their relevance to disease states Describe synthesis of S-adenosylmethionine and its role in methylation reactions Explain how a cobalamin deficiency leads to a secondary folate deficiency Introduction Human body cannot synthesize folic acid Its also called vitamin B9 give rise to tetrahydrofolate (THF), carry one carbon groups ex. Methyl group Intestines releases mostly N5-methy-THF into blood One-carbon (1C) metabolism, mediated by folate cofactor, supports biosynthesis of purines and pyrimidines, aa homeostasis (glycine, serine and methionine) Table 14.4: DM Vasudevan’ s Textbook of Biochemistry for Medical Students 6th edition Enzyme co-factors involved in aa catabolism Involves one of three co-factors: Biotin, Tetrahydrofolate (THF) and S- adenosylmethionine (SAM) These cofactors transfer one-carbon groups in different oxidation states: 1. Biotin transfers carbon in its most oxidized state CO2, it require for catabolism and utilization of branched chain aa • Biotin responsible for carbon dioxide transfer in several carboxylase enzymes Cont-- 2. Tetrahydrofolate (THF) transfers one-carbon groups in intermediate oxidation states and as methyl groups • Tetrahydrobiopterin (BH4, THB) is a cofactor of degradation of phenylalanine • Oxidised form of THF, folate is vitamin for mammals • It converted into THF by DHF reductase 3. S-adenosylmethionine (SAM) transfers methyl groups, most reduced state of carbon THF and SAM imp in aa and nucleotide metabolism SAM used in biosynthesis of creatine, phosphatidylcholine, plasmenylcholine and epinephrine, also methylated DNA, RNA and proteins Enzymes use cobalamin as a cofactor 1. -

Reduction by the Methylreductase System in Methanobacterium Bryantii WILLIAM B
JOURNAL OF BACTERIOLOGY, Jan. 1987, p. 87-92 Vol. 169, No. 1 0021-9193/87/010087-06$02.00/0 Copyright © 1987, American Society for Microbiology Inhibition by Corrins of the ATP-Dependent Activation and CO2 Reduction by the Methylreductase System in Methanobacterium bryantii WILLIAM B. WHITMAN'* AND RALPH S. WOLFE2 Department of Microbiology, University of Georgia, Athens, Georgia 30602,1 and Department of Microbiology, University ofIllinois, Urbana, Illinois 618012 Received 1 August 1986/Accepted 28 September 1986 Corrins inhibited the ATP-dependent activation of the methylreductase system and the methyl coenzyme M-dependent reduction of CO2 in extracts of Methanobacterium bryantii resolved from low-molecular-weight factors. The concentrations of cobinamides and cobamides required for one-half of maximal inhibition of the ATP-depen4ent activation were between 1 and 5 ,M. Cobinamides were more inhibitory at lower concentra- tiops than cobamides. Deoxyadenosylcobalamin was not inhibitory at concentrations up to 25 ,uM. The inhibition of CO2 reduction was competitive with respect to CO2. The concentration of methylcobalamin required for one-half of maximal inhibition was 5 ,M. Other cobamideg inhibited at similar concentrations, but diaquacobinami4e inhibited at lower concentrations. With respect to their affinities and specificities for corrins, inhibition of both the ATP-dependent activation'and CO2 reduction closely resembled the corrin- dependent activation of the methylreductase described in similar extracts (W. B. Whitman and R. S. Wolfe, J. Bacteriol. 164:165-172, 1985). However, whether the multiple effects of corrins are due to action at a single site is unknown. The effect of corrins (cobamides and cobinamides) on in CO2 reduction. -

L-Carnitine, Mecobalamin and Folic Acid Tablets) TRINERVE-LC
For the use of a Registered Medical Practitioner or a Hospital or a Laboratory only (L-Carnitine, Mecobalamin and Folic acid Tablets) TRINERVE-LC 1. Name of the medicinal product Trinerve-LC Tablets 2. Qualitative and quantitative composition Each film- coated tablets contains L-Carnitine…………………….500 mg Mecobalamin……………….1500 mcg Folic acid IP…………………..1.5mg 3. Pharmaceutical form Film- coated tablets 4. Clinical particulars 4.1 Therapeutic indications Vitamin and micronutrient supplementation in the management of chronic disease. 4.2 Posology and method of administration For oral administration only. One tablet daily or as directed by physician. 4.3 Contraindications Hypersensitivity to any constituent of the product. 4.4 Special warnings and precautions for use L-Carnitine The safety and efficacy of oral L-Carnitine has not been evaluated in patients with renal insufficiency. Chronic administration of high doses of oral L-Carnitine in patients with severely compromised renal function or in ESRD patients on dialysis may result in accumulation of the potentially toxic metabolites, trimethylamine (TMA) and trimethylamine-N-oxide (TMAO), since these metabolites are normally excreted in the urine. Mecobalamin Should be given with caution in patients suffering from folate deficiency. The following warnings and precautions suggested with parent form – vitamin B12 The treatment of vitamin B12 deficiency can unmask the symptoms of polycythemia vera. Megaloblastic anemia is sometimes corrected by treatment with vitamin B12. But this can have very serious side effects. Don’t attempt vitamin B12 therapy without close supervision by healthcare provider. Do not take vitamin B12 if Leber’s disease, a hereditary eye disease. -
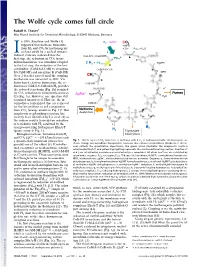
The Wolfe Cycle Comes Full Circle
The Wolfe cycle comes full circle Rudolf K. Thauer1 Max Planck Institute for Terrestrial Microbiology, D-35043 Marburg, Germany n 1988, Rouvière and Wolfe (1) H - ΔμNa+ 2 CO2 suggested that methane formation + MFR from H and CO by methanogenic + 2H+ *Fd + H O I 2 2 ox 2 archaea could be a cyclical process. j O = Indirect evidence indicated that the CoB-SH + CoM-SH fi *Fd 2- a rst step, the reduction of CO2 to for- red R mylmethanofuran, was somehow coupled + * H MPT 2 H2 Fdox 4 to the last step, the reduction of the het- h erodisulfide (CoM-S-S-CoB) to coenzyme CoM-S-S-CoB b MFR M (CoM-SH) and coenzyme B (CoB-SH). H Over 2 decades passed until the coupling C 4 10 mechanism was unraveled in 2011: Via g flavin-based electron bifurcation, the re- CoB-SH duction of CoM-S-S-CoB with H provides 2 H+ the reduced ferredoxin (Fig. 1h) required c + Purines for CO2 reduction to formylmethanofuran ΔμNa + H MPT 4 f H O (2) (Fig. 1a). However, one question still 2 remained unanswered: How are the in- termediates replenished that are removed CoM-SH for the biosynthesis of cell components H Methionine d from CO2 (orange arrows in Fig. 1)? This Acetyl-CoA e anaplerotic (replenishing) reaction has F420 F420H2 recently been identified by Lie et al. (3) as F420 F420H2 the sodium motive force-driven reduction H i of ferredoxin with H2 catalyzed by the i energy-converting hydrogenase EhaA-T H2 (green arrow in Fig. -

Annotation Guidelines for Experimental Procedures
Annotation Guidelines for Experimental Procedures Developed By Mohammed Alliheedi Robert Mercer Version 1 April 14th, 2018 1- Introduction and background information What is rhetorical move? A rhetorical move can be defined as a text fragment that conveys a distinct communicative goal, in other words, a sentence that implies an author’s specific purpose to readers. What are the types of rhetorical moves? There are several types of rhetorical moves. However, we are interested in 4 rhetorical moves that are common in the method section of a scientific article that follows the Introduction Methods Results and Discussion (IMRaD) structure. 1- Description of a method: It is concerned with a sentence(s) that describes experimental events (e.g., “Beads with bound proteins were washed six times (for 10 min under rotation at 4°C) with pulldown buffer and proteins harvested in SDS-sample buffer, separated by SDS-PAGE, and analyzed by autoradiography.” (Ester & Uetz, 2008)). 2- Appeal to authority: It is concerned with a sentence(s) that discusses the use of standard methods, protocols, and procedures. There are two types of this move: - A reference to a well-established “standard” method (e.g., the use of a method like “PCR” or “electrophoresis”). - A reference to a method that was previously described in the literature (e.g., “Protein was determined using fluorescamine assay [41].” (Larsen, Frandesn and Treiman, 2001)). 3- Source of materials: It is concerned with a sentence(s) that lists the source of biological materials that are used in the experiment (e.g., “All microalgal strains used in this study are available at the Elizabeth Aidar Microalgae Culture Collection, Department of Marine Biology, Federal Fluminense University, Brazil.” (Larsen, Frandesn and Treiman, 2001)). -

Untargetted Metabolomic Exploration of the Mycobacterium Tuberculosis Stress Response to Cinnamon Essential Oil
biomolecules Article Untargetted Metabolomic Exploration of the Mycobacterium tuberculosis Stress Response to Cinnamon Essential Oil Elwira Sieniawska 1,* , Rafał Sawicki 2 , Joanna Golus 2 and Milen I. Georgiev 3,4 1 Chair and Department of Pharmacognosy, Medical University of Lublin, Chodzki 1, 20-093 Lublin, Poland 2 Chair and Department of Biochemistry and Biotechnology, Medical University of Lublin, Chodzki 1, 20-093 Lublin, Poland; [email protected] (R.S.); [email protected] (J.G.) 3 Group of Plant Cell Biotechnology and Metabolomics, The Stephan Angeloff Institute of Microbiology, Bulgarian Academy of Sciences, 139 Ruski Blvd., 4000 Plovdiv, Bulgaria; [email protected] 4 Center of Plant Systems Biology and Biotechnology, 4000 Plovdiv, Bulgaria * Correspondence: [email protected] Received: 6 January 2020; Accepted: 24 February 2020; Published: 26 February 2020 Abstract: The antimycobacterial activity of cinnamaldehyde has already been proven for laboratory strains and for clinical isolates. What is more, cinnamaldehyde was shown to threaten the mycobacterial plasma membrane integrity and to activate the stress response system. Following promising applications of metabolomics in drug discovery and development we aimed to explore the mycobacteria response to cinnamaldehyde within cinnamon essential oil treatment by untargeted liquid chromatography–mass spectrometry. The use of predictive metabolite pathway analysis and description of produced lipids enabled the evaluation of the stress symptoms shown by bacteria. This study suggests that bacteria exposed to cinnamaldehyde could reorganize their outer membrane as a physical barrier against stress factors. They probably lowered cell wall permeability and inner membrane fluidity, and possibly redirected carbon flow to store energy in triacylglycerols. Being a reactive compound, cinnamaldehyde may also contribute to disturbances in bacteria redox homeostasis and detoxification mechanisms. -

Downloaded from the Fungene Database (
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.21.307504; this version posted September 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Enhancement of nitrous oxide emissions in soil microbial consortia via copper competition between proteobacterial methanotrophs and denitrifiers Jin Chang,a,b Daehyun Daniel Kim,a Jeremy D. Semrau,b Juyong Lee,a Hokwan Heo,a Wenyu Gu,b* Sukhwan Yoona# aDepartment of Civil and Environmental Engineering, Korea Advanced Institute of Science and Technology, Daejeon, 34141, Korea bDepartment of Civil and Environmental Engineering, University of Michigan, Ann Arbor, MI, 48109 Running Title: Methanotrophic influence on N2O emissions #Address correspondence to Sukhwan Yoon, [email protected]. *Present address: Department of Civil & Environmental Engineering, Stanford University, Palo Alto CA, 94305 bioRxiv preprint doi: https://doi.org/10.1101/2020.09.21.307504; this version posted September 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Unique means of copper scavenging have been identified in proteobacterial methanotrophs, particularly the use of methanobactin, a novel ribosomally synthesized post-translationally modified polypeptide that binds copper with very high affinity. The possibility that copper sequestration strategies of methanotrophs may interfere with copper uptake of denitrifiers in situ and thereby enhance N2O emissions was examined using a suite of laboratory experiments performed with rice paddy microbial consortia. Addition of purified methanobactin from Methylosinus trichosporium OB3b to denitrifying rice paddy soil microbial consortia resulted in substantially increased N2O production, with more pronounced responses observed for soils with lower copper content. -

Enbrace® HR DESCRIPTION: INGREDIENTS
EnBrace® HR with DeltaFolate ™ [1 NF Units] [15 mg DFE Folate] ANTI-ANEMIA PREPARATION as extrinsic/intrinsic factor concentrate plus folate. Prescription Prenatal/Vitamin Drug For Therapeutic Use Multi-phasic Capsules (30ct bottle) NDC 64661-650-30 Rx Only [DRUG] GLUTEN-FREE DESCRIPTION: EnBrace® HR is an orally administered prescription prenatal/vitamin drug for therapeutic use formulated for female macrocytic anemia patients that are in need of treatment, and are under specific direction and monitoring of vitamin B12 and vitamin B9 status by a physician. EnBrace® HR is intended for women of childbearing age who are – or desire to become, pregnant regardless of lactation status. EnBrace® HR may be prescribed for women at risk of depression as a result of folate or cobalamin deficiency - including folate-induced postpartum depression, or are at risk of folate-induced birth defects such as may be found with spina bifida and other neural tube defects (NTDs). INGREDIENTS: Cobalamin intrinsic factor complex 1 NF Units* * National Formulary Units (“NF UNITS”) equivalent to 50 mcg of active coenzyme cobalamin (as cobamamide concentrate with intrinsic factor) ALSO CONTAINS: 1 Folinic acid (B9-vitamer) 2.5 mg + 1 Control-release, citrated folic acid, DHF (B9-Provitamin) 1 mg 2 Levomefolic acid (B9 & B12- cofactor) 5.23 mg 1 6 mg DFE folate (vitamin B9) 2 9 mg DFE l-methylfolate magnesium (molar equivalent). FUNCTIONAL EXCIPIENTS: 13.6 mg FeGC as ferrous glycine cysteinate (1.5 mg 3 3,4 elemental iron ) [colorant], 25 mg ascorbates (24 mg magnesium l-ascorbate, 1 mg zinc l-ascorbate) [antioxidant], at least 23.33 mg phospholipid-omega3 complex5 [marine lipids], 500 mcg betaine (trimethylglycine) [acidifier], 1 mg magnesium l-threonate [stabilizer]. -

5,10-Methylenetetrahydrofolate Dehydrogenase (Ec 1.5.1.5)
Enzymatic Assay of 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE (EC 1.5.1.5) PRINCIPLE: 5,10-MeFH4DH 5,10-Methylene-FH4 + ß-NADP > 5,10-Methenyl-FH4 + ß-NADPH Abbreviations used: 5,10-Methylene-FH4 = 5,10-Methylenetetrahydrofolate 5,10-Methenyl-FH4 = 5,10-Methenyltetrahydrofolate 5,10-MeFH4DH = 5,10-Methylenetetrahydrofolate Dehydrogenase ß-NADP = ß-Nicotinamide Adenine Dinucleotide Phosphate, Oxidized Form ß-NADPH = ß-Nicotinamide Adenine Dinucleotide Phosphate, Reduced Form CONDITIONS: T = 25°C, pH = 7.5, A340nm, Light path = 1 cm METHOD: Continuous Spectrophotometric Rate Determination REAGENTS: A. 50 mM Potassium Phosphate Buffer with 100 mM Potassium Chloride, pH 7.5 at 25°C (Prepare 200 ml in deionized water using Potassium Phosphate, Monobasic, Anhydrous, and Potassium Chloride. Adjust to pH 7.5 at 25°C with 1 M NaOH.) B. 0.002% (w/v) Tetrahydrofolic Acid with 0.002% (v/v) Formaldehyde and 0.1% (v/v) 2-Mercaptoethanol Solution (FH4) (Immediately before used, prepare 100 ml in Reagent A using Tetrahydrofolic Acid, Formaldehyde, 37% Solution,1 and 2-Mercaptoethanol. ) C. 20 mM ß-Nicotinamide Adenine Dinucleotide Phosphate (ß-NADP) (Prepare 2 ml in deionized water using ß-Nicotinamide Adenine Dinucleotide Phosphate, Sodium Salt PREPARE FRESH. ) Page 1 of 3 45-1 Ramsey Road, Shirley, NY 11967, USA Email: [email protected] Tel: 1-631-562-8517 1-631-448-7888 Fax: 1-631-938-8127 Enzymatic Assay of 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE (EC 1.5.1.5) REAGENTS: (continued) D. 5,10-Methylenetetrahydrofolate Dehydrogenase Enzyme Solution (Immediately before use, prepare a solution containing 0.01 - 0.03 unit/ml of 5,10-Methylenetetrahydrofolate Dehydrogenase in cold Reagent A.) PROCEDURE: Pipette (in milliliters) the following reagents into suitable cuvettes: Test Blank Reagent B (FH4) 2.80 2.80 Reagent D (Enzyme Solution) 0.10 0.10 Mix by inversion and equilibrate to 25°C. -

Widespread Soil Bacterium That Oxidizes Atmospheric Methane
Widespread soil bacterium that oxidizes atmospheric methane Alexander T. Tveita,1, Anne Grethe Hestnesa,1, Serina L. Robinsona, Arno Schintlmeisterb, Svetlana N. Dedyshc, Nico Jehmlichd, Martin von Bergend,e, Craig Herboldb, Michael Wagnerb, Andreas Richterf, and Mette M. Svenninga,2 aDepartment of Arctic and Marine Biology, Faculty of Biosciences, Fisheries and Economics, UiT The Arctic University of Norway, 9037 Tromsoe, Norway; bCenter of Microbiology and Environmental Systems Science, Division of Microbial Ecology, University of Vienna, 1090 Vienna, Austria; cWinogradsky Institute of Microbiology, Research Center of Biotechnology of Russian Academy of Sciences, 117312 Moscow, Russia; dDepartment of Molecular Systems Biology, Helmholtz Centre for Environmental Research-UFZ, 04318 Leipzig, Germany; eFaculty of Life Sciences, Institute of Biochemistry, University of Leipzig, 04109 Leipzig, Germany; and fCenter of Microbiology and Environmental Systems Science, Division of Terrestrial Ecosystem Research, University of Vienna, 1090 Vienna, Austria Edited by Mary E. Lidstrom, University of Washington, Seattle, WA, and approved March 7, 2019 (received for review October 22, 2018) The global atmospheric level of methane (CH4), the second most as-yet-uncultured clades within the Alpha- and Gammaproteobacteria important greenhouse gas, is currently increasing by ∼10 million (16–18) which were designated as upland soil clusters α and γ tons per year. Microbial oxidation in unsaturated soils is the only (USCα and USCγ, respectively). Interest in soil atmMOB has known biological process that removes CH4 from the atmosphere, increased significantly since then because they are responsible but so far, bacteria that can grow on atmospheric CH4 have eluded for the only known biological removal of atmospheric CH4 all cultivation efforts. -

Global Molecular Analyses of Methane Metabolism in Methanotrophic Alphaproteobacterium, Methylosinus Trichosporium Ob3b
ORIGINAL RESEARCH ARTICLE published: 03 April 2013 doi: 10.3389/fmicb.2013.00070 Global molecular analyses of methane metabolism in methanotrophic Alphaproteobacterium, Methylosinus trichosporium OB3b. Part II. metabolomics and 13C-labeling study SongYang 1, Janet B. Matsen1, Michael Konopka1, Abigail Green-Saxena2, Justin Clubb1, Martin Sadilek 3, Victoria J. Orphan4, David Beck 1,5 and Marina G. Kalyuzhnaya6* 1 Department of Chemical Engineering, University of Washington, Seattle, WA, USA 2 Division of Biology, California Institute of Technology, Pasadena, CA, USA 3 Department of Chemistry, University of Washington, Seattle, WA, USA 4 Division of Geological and Planetary Sciences; California Institute of Technology, Pasadena, CA, USA 5 eScience Institute, University of Washington, Seattle, WA, USA 6 Department of Microbiology, University of Washington, Seattle, WA, USA Edited by: In this work we use metabolomics and 13C-labeling data to refine central metabolic Amy V. Callaghan, University of pathways for methane utilization in Methylosinus trichosporium OB3b, a model alphapro- Oklahoma, USA teobacterial methanotrophic bacterium. We demonstrate here that similar to non-methane Reviewed by: Alan A. DiSpirito, Ohio State utilizing methylotrophic alphaproteobacteria the core metabolism of the microbe is rep- University, USA resented by several tightly connected metabolic cycles, such as the serine pathway, the Amy V. Callaghan, University of ethylmalonyl-CoA (EMC) pathway, and the citric acid (TCA) cycle. Both in silico estimations Oklahoma, USA and stable isotope labeling experiments combined with single cell (NanoSIMS) and bulk *Correspondence: biomass analyses indicate that a significantly larger portion of the cell carbon (over 60%) is Marina G. Kalyuzhnaya, Department 13 of Microbiology, University of derived from CO2 in this methanotroph.