Table S1. List of Dendrobatoid Samples Included in Our Phylogenomic Analyses and Associated Locality Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Article Look but Don't Lick! Find out How These Brightly Colored Frogs
The Power of Poison Look But Don’t Lick! GRADES K-2 NOTES FOR EDUCATORS Common Core State Standards: W.K-2.2, W.K-2.8 Have students read the article “Look but Don’t Lick!” Have them write notes RI.K-2.1, RI.K-2.2, RI.K-2.4, RI.K-2.7, RI.K-2.10 in the large right-hand margin. For example, they could underline key passages, paraphrase important information, or write down questions that New York State Science Core Curriculum: they have. LE 3.1a Next Generation Science Standards: If it is not possible to create color handouts, use a computer projector to PE 1-LS1-2 display the reading so that students can see the colorful frog photos. You DCI LS1.A: Structure and Function may also have them color their black and white copies to match the actual All organisms have external parts. Different colors. animals use their body parts in different ways to see, hear, grasp objects, protect Ask: themselves, move from place to place, and • What does it mean for an animal to be poisonous? (A: An animal is seek, find, and take in food, water and air. poisonous if its body contains a substance that is harmful or fatal to other Plants also have different parts (roots, animals.) stems, leaves, flowers, fruits) that help • How does being poisonous help the frogs in this article survive? (A: them survive and grow. Predators will not eat an animal that is poisonous to them. The frogs signal that they are poisonous by their bright colors, which warn predators not to eat them. -

Universidade Vila Velha Programa De Pós-Graduação Em Ecologia De Ecossistemas
UNIVERSIDADE VILA VELHA PROGRAMA DE PÓS-GRADUAÇÃO EM ECOLOGIA DE ECOSSISTEMAS MODELOS DE NICHO ECOLÓGICO E A DISTRIBUIÇÃO DE PHYLLODYTES (ANURA, HYLIDAE): UMA PERSPECTIVA TEMPORAL DE UM GÊNERO POTENCIALMENTE AMEAÇADO DE EXTINÇÃO POR MUDANÇAS CLIMÁTICAS E INTERAÇÕES BIOLÓGICAS MARCIO MAGESKI MARQUES VILA VELHA FEVEREIRO / 2018 UNIVERSIDADE VILA VELHA PROGRAMA DE PÓS-GRADUAÇÃO EM ECOLOGIA DE ECOSSISTEMAS MODELOS DE NICHO ECOLÓGICO E A DISTRIBUIÇÃO DE PHYLLODYTES (ANURA, HYLIDAE): UMA PERSPECTIVA TEMPORAL DE UM GÊNERO POTENCIALMENTE AMEAÇADO DE EXTINÇÃO POR MUDANÇAS CLIMÁTICAS E INTERAÇÕES BIOLÓGICAS Tese apresentada a Universidade Vila Velha, como pré-requisito do Programa de Pós- Graduação em Ecologia de Ecossistemas, para obtenção do título de Doutor em Ecologia. MARCIO MAGESKI MARQUES VILA VELHA FEVEREIRO / 2018 À minha esposa Mariana e meu filho Ângelo pelo apoio incondicional em todos os momentos, principalmente nos de incerteza, muito comuns para quem tenta trilhar novos caminhos. AGRADECIMENTOS Seria impossível cumprir essa etapa tão importante sem a presença do divino Espírito Santo de Deus, de Maria Santíssima dos Anjos e Santos. Obrigado por me fortalecerem, me levantarem e me animarem diante das dificultades, que foram muitas durante esses quatro anos. Agora, servirei a meu Deus em mais uma nova missão. Muito Obrigado. À minha amada esposa Mariana que me compreendeu e sempre esteve comigo me apoiando durante esses quatro anos (na verdade seis, se contar com o mestrado) em momentos de felicidades, tristezas, ansiedade, nervosismo, etc... Esse período nos serviu para demonstrar o quanto é forte nosso abençoado amor. Sem você isso não seria real. Te amo e muito obrigado. Ao meu amado filho, Ângelo Miguel, que sempre me recebia com um iluminado sorriso e um beijinho a cada vez que eu chegava em casa depois de um dia de trabalho. -

Anura: Dendrobatidae) EN EL BOSQUE PLUVIAL TROPICAL EN EL DEPARTAMENTO DEL CHOCÓ, COLOMBIA*
BOLETÍN CIENTÍFICO bol.cient.mus.hist.nat. 23 (1), enero-junio, 2019. 85-97. ISSN: 0123-3068 (Impreso) ISSN: 2462-8190 (En línea) CENTRO DE MUSEOS MUSEO DE HISTORIA NATURAL COMPOSICIÓN DIETARIA DE DOS ESPECIES DEL GÉNERO Andinobates (Anura: Dendrobatidae) EN EL BOSQUE PLUVIAL TROPICAL EN EL DEPARTAMENTO DEL CHOCÓ, COLOMBIA* Lucellis Maria Rivas1, Jhon Brayan García2, Jhon Tailor Rengifo3 Resumen Objetivo: Determinar la composición dietaría de Andinobates en zonas de bosque pluvial tropical en el departamento del Chocó, Colombia, para incrementar el conocimiento de los Dendrobatidae en la región pacifica colombiana. Metodología: Se utilizó el método de relevamiento por encuentros visuales (VES); además se calculó el índice de importancia relativa, el índice de jerarquización y la diversidad trófica. Resultados: Se identificaron de acuerdo a la nomenclatura taxonómica un total de 140 individuos repartidos en dos phylum, tres clases y cinco ordenes, siendo el orden Hy- menoptera el más abundante para las dos especies; por otro lado, no se predijeron más novedades tróficas. Conclusiones: Los índices permiten inferir que las especies estudiadas consumen una amplia variedad de invertebrados, por ello estas especies pueden ser especialistas o generalistas. Palabras clave: dieta, Chocó, diversidad trófica, contenidos estomacales Hymenoptera. DIETARY COMPOSITION OF TWO SPECIES OF THE GENRE Andinobates (Anura: Dendrobatidae) IN THE TROPICAL PLUVIAL FOREST IN THE CHOCÓ DEPARTMENT, COLOMBIA Abstract Objectives: To determine the dietary composition of Andinobates in tropical rain forest areas in the Department of Chocó, Colombia, to increase the knowledge of dendrobatids in the Colombian Pacific region. Methodology: The visual encounter survey method (VES) was used. In addition, the relative importance index, the hierarchy index, and the trophic diversity were calculated. -

The Tadpole of Adelphobates Galactonotus (Steindachner, 1864) (Amphibia, Anura, Dendrobatidae)
Zootaxa 4422 (2): 287–290 ISSN 1175-5326 (print edition) http://www.mapress.com/j/zt/ Correspondence ZOOTAXA Copyright © 2018 Magnolia Press ISSN 1175-5334 (online edition) https://doi.org/10.11646/zootaxa.4422.2.8 http://zoobank.org/urn:lsid:zoobank.org:pub:C16E5D1D-D75C-4411-94AE-41C038CA7BA9 The tadpole of Adelphobates galactonotus (Steindachner, 1864) (Amphibia, Anura, Dendrobatidae) DANUSY LOPES SANTOS1,2, 3, SILIONAMÃ PEREIRA DANTAS4 & FAUSTO NOMURA1 1Laboratório de Herpetologia e Comportamento Animal, Departamento de Ecologia, Universidade Federal de Goiás. Campus Samambaia, CEP 74001-970, Goiânia, GO, Brazil 2Laboratório de Ecologia Teórica, Departamento de Zoologia e Botânica, Universidade Estadual Paulista, Campus São José do Rio Preto. CEP 15054-000, São José do Rio Preto, São Paulo, Brazil 4Universidade Federal de Tocantins, Campus Araguaína, CEP 77838-824, Araguaína, TO, Brazil 3Corresponding author. E-mail: [email protected] The systematics of the dart-poison frogs, family Dendrobatidae, experienced several taxonomic rearrangements over time (e.g., Grant et al. 2006, 2017; Brown et al. 2011). Currently, this family comprises 194 described species organized in three sub-families and 15 genera (Frost 2018). Among them, the genus Adelphobates Grant, Frost, Caldwell, Gagliardo, Haddad, Kok, Means, Noonan, Schargel, & Wheeler, 2006, comprises three species, all distributed in Central and lower Amazon drainage of Peru and Brazil, and possibly in northeast of Bolivia (Grant et al. 2006; Frost 2018). Adelphobates galactonotus (Steindachner 1864) is an endemic Brazilian frog, and can be found throughout Pará, Maranhão, Mato Grosso and Tocantins states (Hoogmoed & Avila-Pires 2012), related to Amazon forest formations and also in transitional areas between the Cerrado and the Amazon forest (Valdujo et al. -

Chromosome Analysis of Five Brazilian
c Indian Academy of Sciences RESEARCH ARTICLE Chromosome analysis of five Brazilian species of poison frogs (Anura: Dendrobatidae) PAULA CAMARGO RODRIGUES1, ODAIR AGUIAR2, FLÁVIA SERPIERI1, ALBERTINA PIMENTEL LIMA3, MASAO UETANEBARO4 and SHIRLEI MARIA RECCO-PIMENTEL1∗ 1Departamento de Anatomia, Biologia Celular e Fisiologia, Instituto de Biologia, Universidade Estadual de Campinas, 13083-863 Campinas, São Paulo, Brazil 2Departamento de Biociências, Universidade Federal de São Paulo, Campus Baixada Santista, 11060-001 Santos, São Paulo, Brazil 3Coordenadoria de Pesquisas em Ecologia, Instituto Nacional de Pesquisas do Amazonas, 69011-970 Manaus, Amazonas, Brazil 4Departamento de Biologia, Universidade Federal de Mato Grosso do Sul, 70070-900 Campo Grande, Mato Grosso do Sul, Brazil Abstract Dendrobatid frogs have undergone an extensive systematic reorganization based on recent molecular findings. The present work describes karyotypes of the Brazilian species Adelphobates castaneoticus, A. quinquevittatus, Ameerega picta, A. galactonotus and Dendrobates tinctorius which were compared to each other and with previously described related species. All karyotypes consisted of 2n = 18 chromosomes, except for A. picta which had 2n = 24. The karyotypes of the Adelphobates and D. tinctorius species were highly similar to each other and to the other 2n = 18 previously studied species, revealing conserved karyotypic characteristics in both genera. In recent phylogenetic studies, all Adelphobates species were grouped in a clade separated from the Dendrobates species. Thus, we hypothesized that their common karyotypic traits may have a distinct origin by chromosome rearrangements and mutations. In A. picta, with 2n = 24, chromosome features of pairs from 1 to 8 are shared with other previously karyotyped species within this genus. Hence, the A. -
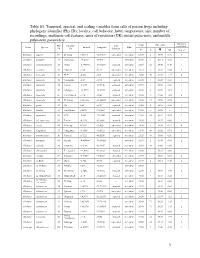
1 Table S1. Temporal, Spectral, and Scaling Variables from Calls Of
Table S1. Temporal, spectral, and scaling variables from calls of poison frogs including phylogeny identifier (Phy ID), locality, call behavior, habit, temperature, size, number of recordings, multinote call features, units of repetition (UR), initial pulse-note, and middle pulse-note parameters. Analyzed Phy Locality Call Temp SVL (mm) Genus Species Latitude Longitude Habit recordings ID ID Behavior °C N ! SD N of ♂ Allobates algorei 60 El Tama 7.65375 -72.19137 concealed terrestrial 23.50 8 18.90 0.70 3 Allobates brunneus 37 Guimaraes -15.2667 -55.5311 -- terrestrial 26.50 1 16.13 0.00 1 Allobates caeruleodactylus 48 Borba -4.398593 -59.60251 exposed terrestrial 25.60 12 15.50 0.40 1 Allobates crombiei 52 Altamira -3.65 -52.38 concealed terrestrial 24.10 2 18.10 0.04 2 Allobates femoralis 43 ECY -0.633 -76.5 concealed terrestrial 25.60 20 23.58 1.27 6 Allobates femoralis 46 Porongaba -8.67 -72.78 exposed terrestrial 25.00 1 25.38 0.00 1 Allobates femoralis 44 Leticia -4.2153 -69.9406 exposed terrestrial 25.50 1 20.90 0.00 1 Allobates femoralis 40 Albergue -12.8773 -71.3865 exposed terrestrial 26.00 6 21.98 2.18 1 Allobates femoralis 41 CAmazonico -12.6 -70.08 exposed terrestrial 26.00 12 22.43 1.06 4 Allobates femoralis 45 El Palmar 8.333333 -61.66667 concealed terrestrial 24.00 27 25.50 0.76 1 Allobates granti 49 FG 3.62 -53.17 exposed terrestrial 24.60 8 16.15 0.55 1 Allobates humilis 59 San Ramon 8.8678 -70.4861 concealed terrestrial 19.50 -- 21.80 -- 1 Allobates insperatus 54 ECY -0.633 -76.4005 exposed terrestrial 24.60 18 16.64 0.93 7 Allobates aff. -

Visual Signaling in Anuran Amphibians
.. Hödl, W. and Amezquita, A. (2001). Visual signaling in anuran amphibians. In: Anuran communication, (M.J. Ryan, ed.). .. Smithsonian lust. Press, Washington. Pp. 121-141. 10 WALTER HÖDL AND ADOLFO AMEZQUITA Visual Signaling in Anuran Amphibians lntroduction cation. social behavior, or natural history. visual signaling was either not considered or was treated as a minor subject Acoustic communication plays a fundamental role in an- (Wells 1977a, 1977b; Arak 1983; Duellman and Trueb 1986; uran reproduction and thus is involved in evolutionary Rand 1988; Halliday and Tejedo 1995; Stebbins and Cohen processes such as mate recognition. reproductive isolation. 1995; Sullivan et al. 1995). The most detailed review ofthe speciation. and character displacement (Wells 1977a. 1977b. subject is now more than 20 years old (Wells 1977b). Never- 1988;Rand 1988;Gerhardt and Schwartz 1995;Halliday and theless some authors have discussed the possible evolution- Tejedo 1995;Sullivan et al. 1995).Visual cues. however. have ary link between visual signaling and the reproductive ecol- been thought to function only during dose-range inter- ogy of species, such as reproduction associated with streams actions (Wells 1977c; Duellman and Trueb 1986). Visual sig- (Heyer et aI. 1990; Lindquist and Hetherington 1996. 1998; naling is predicted to be predominantly employed by diur- Hödl et al. 1997;Haddad and Giaretta 1999) or reproduction nal species at sites with an unobstructed view (Endler 1992). within feeding territories (Wells 1977c). Diurnality. however. is not common for the majority offrog Our aim in this review is (1) to propose a dassmcation of species. Thus vocalizations. which are highly efficient for reported behavioral patterns of visual signaling in frags; (2) communicating at night or in dense vegetation, are by far to describe the diversity of visual signals among living an- the best studied anuran signals (Duellman and Trueb 1986; uran taxa; and (3) to apply a comparative approach to explor- Fritzsch et aI. -

A Review of Chemical Defense in Poison Frogs (Dendrobatidae): Ecology, Pharmacokinetics, and Autoresistance
Chapter 21 A Review of Chemical Defense in Poison Frogs (Dendrobatidae): Ecology, Pharmacokinetics, and Autoresistance Juan C. Santos , Rebecca D. Tarvin , and Lauren A. O’Connell 21.1 Introduction Chemical defense has evolved multiple times in nearly every major group of life, from snakes and insects to bacteria and plants (Mebs 2002 ). However, among land vertebrates, chemical defenses are restricted to a few monophyletic groups (i.e., clades). Most of these are amphibians and snakes, but a few rare origins (e.g., Pitohui birds) have stimulated research on acquired chemical defenses (Dumbacher et al. 1992 ). Selective pressures that lead to defense are usually associated with an organ- ism’s limited ability to escape predation or conspicuous behaviors and phenotypes that increase detectability by predators (e.g., diurnality or mating calls) (Speed and Ruxton 2005 ). Defended organisms frequently evolve warning signals to advertise their defense, a phenomenon known as aposematism (Mappes et al. 2005 ). Warning signals such as conspicuous coloration unambiguously inform predators that there will be a substantial cost if they proceed with attack or consumption of the defended prey (Mappes et al. 2005 ). However, aposematism is likely more complex than the simple pairing of signal and defense, encompassing a series of traits (i.e., the apose- matic syndrome) that alter morphology, physiology, and behavior (Mappes and J. C. Santos (*) Department of Zoology, Biodiversity Research Centre , University of British Columbia , #4200-6270 University Blvd , Vancouver , BC , Canada , V6T 1Z4 e-mail: [email protected] R. D. Tarvin University of Texas at Austin , 2415 Speedway Stop C0990 , Austin , TX 78712 , USA e-mail: [email protected] L. -

Taxonomic Checklist of Amphibian Species Listed in the CITES
CoP17 Doc. 81.1 Annex 5 (English only / Únicamente en inglés / Seulement en anglais) Taxonomic Checklist of Amphibian Species listed in the CITES Appendices and the Annexes of EC Regulation 338/97 Species information extracted from FROST, D. R. (2015) "Amphibian Species of the World, an online Reference" V. 6.0 (as of May 2015) Copyright © 1998-2015, Darrel Frost and TheAmericanMuseum of Natural History. All Rights Reserved. Additional comments included by the Nomenclature Specialist of the CITES Animals Committee (indicated by "NC comment") Reproduction for commercial purposes prohibited. CoP17 Doc. 81.1 Annex 5 - p. 1 Amphibian Species covered by this Checklist listed by listed by CITES EC- as well as Family Species Regulation EC 338/97 Regulation only 338/97 ANURA Aromobatidae Allobates femoralis X Aromobatidae Allobates hodli X Aromobatidae Allobates myersi X Aromobatidae Allobates zaparo X Aromobatidae Anomaloglossus rufulus X Bufonidae Altiphrynoides malcolmi X Bufonidae Altiphrynoides osgoodi X Bufonidae Amietophrynus channingi X Bufonidae Amietophrynus superciliaris X Bufonidae Atelopus zeteki X Bufonidae Incilius periglenes X Bufonidae Nectophrynoides asperginis X Bufonidae Nectophrynoides cryptus X Bufonidae Nectophrynoides frontierei X Bufonidae Nectophrynoides laevis X Bufonidae Nectophrynoides laticeps X Bufonidae Nectophrynoides minutus X Bufonidae Nectophrynoides paulae X Bufonidae Nectophrynoides poyntoni X Bufonidae Nectophrynoides pseudotornieri X Bufonidae Nectophrynoides tornieri X Bufonidae Nectophrynoides vestergaardi -

F3999f15-C572-46Ad-Bbbe
THE STATUTES OF THE REPUBLIC OF SINGAPORE ENDANGERED SPECIES (IMPORT AND EXPORT) ACT (CHAPTER 92A) (Original Enactment: Act 5 of 2006) REVISED EDITION 2008 (1st January 2008) Prepared and Published by THE LAW REVISION COMMISSION UNDER THE AUTHORITY OF THE REVISED EDITION OF THE LAWS ACT (CHAPTER 275) Informal Consolidation – version in force from 22/6/2021 CHAPTER 92A 2008 Ed. Endangered Species (Import and Export) Act ARRANGEMENT OF SECTIONS PART I PRELIMINARY Section 1. Short title 2. Interpretation 3. Appointment of Director-General and authorised officers PART II CONTROL OF IMPORT, EXPORT, ETC., OF SCHEDULED SPECIES 4. Restriction on import, export, etc., of scheduled species 5. Control of scheduled species in transit 6. Defence to offence under section 4 or 5 7. Issue of permit 8. Cancellation of permit PART III ENFORCEMENT POWERS AND PROCEEDINGS 9. Power of inspection 10. Power to investigate and require information 11. Power of entry, search and seizure 12. Powers ancillary to inspections and searches 13. Power to require scheduled species to be marked, etc. 14. Power of arrest 15. Forfeiture 16. Obstruction 17. Penalty for false declarations, etc. 18. General penalty 19. Abetment of offences 20. Offences by bodies corporate, etc. 1 Informal Consolidation – version in force from 22/6/2021 Endangered Species (Import and 2008 Ed. Export) CAP. 92A 2 PART IV MISCELLANEOUS Section 21. Advisory Committee 22. Fees, etc., payable to Board 23. Board not liable for damage caused to goods or property as result of search, etc. 24. Jurisdiction of court, etc. 25. Composition of offences 26. Exemption 27. Service of documents 28. -

Zootaxa, Revision of the Ranitomeya Fantastica Species Complex with Description Of
TERM OF USE This pdf is provided by Magnolia Press for private/research use. Commercial sale or deposition in a public library or website site is prohibited. Zootaxa 1823: 1–24 (2008) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ ZOOTAXA Copyright © 2008 · Magnolia Press ISSN 1175-5334 (online edition) Revision of the Ranitomeya fantastica species complex with description of two new species from Central Peru (Anura: Dendrobatidae) JASON L. BROWN1,4, EVAN TWOMEY1,5, MARK PEPPER2 & MANUEL SANCHEZ RODRIGUEZ3 1Department of Biology, East Carolina University, Greenville, NC, USA 2Understory Enterprises, Charing Cross, Ontario, Canada 3Understory Enterprises, Iquitos, Peru 4Corresponding author. E-mail: [email protected] 5Corresponding author. E-mail:[email protected] Abstract We describe two new species of poison frogs (genus Ranitomeya) from the central Rio Huallaga drainage and adjacent Cordillera Azul in central Peru. Both species were previously considered to be members of Ranitomeya fantastica, a spe- cies described from the town of Yurimaguas, Peru. Extensive sampling of putative R. fantastica (including near-topo- typic material) throughout central Peru, and the resulting morphological and phylogenetic analysis has led us to conclude that R. fantastica sensu lato is a complex of three closely related species rather than a single, widely distributed species. The first of these species occurs near the type locality of R. fantastica but bears significant dissimilarity to the original type series and forms a monophyletic clade that is distributed throughout an expansive lowland zone between Rio Huall- aga and Rio Ucayali. This species is diagnosable by its brilliant red head and advertisement call differences. -

Silverstone, 1976
A REVISION OF THE POISON-ARROW FROGS OF THE GENUS PHYLLOBATES BIBRON IN SAGRA (FAMILY DENDROBATIDAE) By PHILIP A. SILVERSTOi'E NATURAL HISTORY M US EU ~ l OF LOS ANGELES COUNTY SCIEl'CE BULLETI!" ~ 7 • :-,:ovEMBER 25. 197b A REVISIO:\, OF THE POISO:\'-ARRO\\' FROGS OF THE GE:\'US PHYUOBATES BIBRO:-': IS SAGRA ( FA ~ lI lY DE:-':DROBATIDAE) A REVISION OF THE POISON-ARROW FROGS OF THE GENUS PHYLLOBATES BIBRON IN SAGRA (FAMILY DENDROBATIDAE) By PHILIP A. SILVERSTONE :-<ATU RAL HISTORY :\1U SElJ~ l OF LOS AV iELES COUI\TY SCIE!\CE BULLETl I\ 17 • :-;OVBlB ER ~5. 1976 T AB LE OF CON TE NTS AC,,"OWLEDGMESTS ~h T U I " LS ""D ~l ET HOOS ~ u-e um abbrevialion$ 3 PNlUJJA< 1a BIB_ ,.,S..u...1841 J Ddinilion J D i a ~ nos i s J Descripelon J Conten t , Tad poles , Specie, group' 7 Life history 7 Food , Summary of distribution 9 Key 10 the genera of Dendrobatidac 9 Key 10 the species of Ph.l'lIoh/ll'-f 9 8 K."OW ~ (ilt 0\Jl' 20 Phylloha tt!_, ,pec ies 20 PhylloM tt!s aurorut!nio 21 Phyllooott!Sbiro /or 2J Phylloha lt!S lugubris 24 Phy/foha tt!s ,';IlOlll.S zs F EMoifAUS (iltOliP 27 PhylJoM u s a'llltwly'; 27 Ph)·fJobou s bol'lt!ngt!ri •• aa Phy/Jobatu t!spino so; 29 Phy/Jobatt!Sjt!mo ralis 50 Phyllobau s tricohv J2 Ph,.llobau s zapar o JJ P O f!S (; 10l/P' as Phyllobatt!s bolMu lllU Phyllooolt!S ingt!r; " Phyllohult' J pan'ulus "J6 Phyllobalt!S peters! J7 Phyllohules plctu s aa Phvltobates pulrhripeclu .f 4J PhyIJob/lles ,marulld;nus 44 T~ lVm A I /JS (; IO\; P Phyllobalt!s bault! ri " Ph.l'lIobalt!s triviuOl,.s " " U TElt U l,IRE CnED 50 FRONTI,PII:n: II.