Linkage and Crossing Over
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetic Effects on Microsatellite Diversity in Wild Emmer Wheat (Triticum Dicoccoides) at the Yehudiyya Microsite, Israel
Heredity (2003) 90, 150–156 & 2003 Nature Publishing Group All rights reserved 0018-067X/03 $25.00 www.nature.com/hdy Genetic effects on microsatellite diversity in wild emmer wheat (Triticum dicoccoides) at the Yehudiyya microsite, Israel Y-C Li1,3, T Fahima1,MSRo¨der2, VM Kirzhner1, A Beiles1, AB Korol1 and E Nevo1 1Institute of Evolution, University of Haifa, Mount Carmel, Haifa 31905, Israel; 2Institute for Plant Genetics and Crop Plant Research, Corrensstrasse 3, 06466 Gatersleben, Germany This study investigated allele size constraints and clustering, diversity. Genome B appeared to have a larger average and genetic effects on microsatellite (simple sequence repeat number (ARN), but lower variance in repeat number 2 repeat, SSR) diversity at 28 loci comprising seven types of (sARN), and smaller number of alleles per locus than genome tandem repeated dinucleotide motifs in a natural population A. SSRs with compound motifs showed larger ARN than of wild emmer wheat, Triticum dicoccoides, from a shade vs those with perfect motifs. The effects of replication slippage sun microsite in Yehudiyya, northeast of the Sea of Galilee, and recombinational effects (eg, unequal crossing over) on Israel. It was found that allele distribution at SSR loci is SSR diversity varied with SSR motifs. Ecological stresses clustered and constrained with lower or higher boundary. (sun vs shade) may affect mutational mechanisms, influen- This may imply that SSR have functional significance and cing the level of SSR diversity by both processes. natural constraints. -

Biology Dictionary English - Khmer
vcnanuRkm CIvviTüa Gg;eKøs-Exµr Biology Dictionary English - Khmer saklviTüal½yPUminÞPñMeBj ed)a:tWm:g; CIvviTüa e)aHBum<elIkTI 3 ¬EksMrYl¦ 2003 Preface to the Third Edition (Revised) This dictionary is the work of many teachers and some students in the Biology department of The Royal University of Phnom Penh. It has developed over the last three years in response to the need of Biology students to learn Biology from English text books. We have also tried to anticipate the future needs of Biology students and teachers in Cambodia. If they want to join the global scientific community; read scientific journals, listen to international media, attend international conferences or study outside Cambodia, then they will probably need to communicate in English. Therefore, the main aim of this book is to help Cambodian students and teachers at the university level to understand Biology in English. All languages evolve. In the past the main influence on Khmer language was French. Nowadays, it is increasingly English. Some technical terms have already been absorbed from French and have become Khmer. Nowadays new technical terms are usually created in English and are used around the world. Language is also created by those who use it and only exists when it is used. Therefore, common usage has also influenced our translation. We have tried to respond to these various influences when preparing this dictionary, so that it represents many different opinions - old and new, Francophile, Anglophile and Khmer. But there will always be some disagreement about the translation of some terms. This is normal and occurs in all languages. -

Molecular Biology and Applied Genetics
MOLECULAR BIOLOGY AND APPLIED GENETICS FOR Medical Laboratory Technology Students Upgraded Lecture Note Series Mohammed Awole Adem Jimma University MOLECULAR BIOLOGY AND APPLIED GENETICS For Medical Laboratory Technician Students Lecture Note Series Mohammed Awole Adem Upgraded - 2006 In collaboration with The Carter Center (EPHTI) and The Federal Democratic Republic of Ethiopia Ministry of Education and Ministry of Health Jimma University PREFACE The problem faced today in the learning and teaching of Applied Genetics and Molecular Biology for laboratory technologists in universities, colleges andhealth institutions primarily from the unavailability of textbooks that focus on the needs of Ethiopian students. This lecture note has been prepared with the primary aim of alleviating the problems encountered in the teaching of Medical Applied Genetics and Molecular Biology course and in minimizing discrepancies prevailing among the different teaching and training health institutions. It can also be used in teaching any introductory course on medical Applied Genetics and Molecular Biology and as a reference material. This lecture note is specifically designed for medical laboratory technologists, and includes only those areas of molecular cell biology and Applied Genetics relevant to degree-level understanding of modern laboratory technology. Since genetics is prerequisite course to molecular biology, the lecture note starts with Genetics i followed by Molecular Biology. It provides students with molecular background to enable them to understand and critically analyze recent advances in laboratory sciences. Finally, it contains a glossary, which summarizes important terminologies used in the text. Each chapter begins by specific learning objectives and at the end of each chapter review questions are also included. -

The Importance of Genetic Redundancy in Evolution
TREE 2688 No. of Pages 14 Trends in Ecology & Evolution Review The Importance of Genetic Redundancy in Evolution Áki J. Láruson,1,3,*,@ Sam Yeaman,2,@ and Katie E. Lotterhos1,3,@ Genetic redundancy has been defined in many different ways at different levels Highlights of biological organization. Here, we briefly review the general concept of redun- The use of the term ‘genetic redundancy’ dancy and focus on the evolutionary importance of redundancy in terms of the has changed over the past 100 years. number of genotypes that give rise to the same phenotype. We discuss the chal- The amount of redundancy in the lenges in determining redundancy empirically, with published experimental mapping of genotype to phenotype is a examples, and demonstrate the use of the C-score metric to quantify redun- critical parameter for evolutionary out- dancy in evolution studies. We contrast the implicit assumptions of redundancy comes under a range of models. in quantitative versus population genetic models, show how this contributes to Distinguishing the conceptual terms signatures of allele frequency shifts, and highlight how the rapid accumulation ‘genotypic redundancy’ and ‘segregating of genome-wide association data provides an avenue for further understanding redundancy’ promotes clearer language the prevalence and role of redundancy in evolution. to further the study of redundancy at all levels of evolutionary biology. History of Redundancy in Evolutionary Studies Empirical determination of redundancy is Redundancy describes a situation in which there is an excess of causal components in a system, challenging, but there are approaches above the minimum needed for its proper function. Understanding how redundancy is built into which allow for the quantitative inference of redundancy. -

Recombination and the Evolution of Satellite DNA
Genet. Res., Comb. (1986), 47, pp. 167-174 With 1 text-figure Printed in Great Britain 167 Recombination and the evolution of satellite DNA WOLFGANG STEPHAN* Institute of Animal Genetics, University of Edinburgh, West Mains Road, Edinburgh EH9 3JN, U.K. (Received 11 September 1985 and in revised form 6 December 1985) Summary In eukaryotic chromosomes, large blocks of satellite DNA are associated with regions of reduced meiotic recombination. No function of highly repeated, tandemly arranged DNA sequences has been identified so far at the cellular level, though the structural properties of satellite DNA are relatively well known. In studying the joint action of meiotic recombination, genetic drift and natural selection on the copy number of a family of highly repeated DNA (HRDNA), this paper looks at the structure-function debate for satellite DNA from the standpoint of molecular population genetics. It is shown that (i) HRDNA accumulates most probably in regions of near zero crossing over (heterochromatin), and that (ii), due to random genetic drift the effect of unequal crossover on copy numbers is stronger, the smaller the population size. As a consequence, highly repeated sequences are likely to persist longest (over evolutionary times) in small populations. The results are based on a fairly general class of models of unequal crossing over and natural selection which have been treated both analytically and by computer simulation. 1. Introduction lem of the function of these DNAs has been the sub- Eukaryotic chromosomes contain nucleotide se- ject of major controversies during the past two quences of lengths from about 10 to several hundred decades. -

Laboratory III Linkage
Genetics Laboratory III Biology 303 Spring 2007 Linkage Dr. Wadsworth Introduction same chromosome, the meiotic assortment of chromosomes alone would not predict the Genes are arranged as units in a linear order independent assortment of genes on those along chromosomes. The position of each gene chromosomes. In the absence of gene along the chromosome is called its locus. Each assortment, we would predict that the progeny gene has a unique chromosomal position and could only have the allelic combinations that therefore a unique locus. In fact, geneticists were present in the parent chromosomes. Genes often use the term genetic locus interchangeably that do not assort are said to show complete with the term gene. linkage. Alternatively, genes that independently assort are said to be unlinked. For the following The relative position of genes along a two examples, compare the genotypes and chromsomes can be analyzed by determining phenotypes generated in a dihybrid crosses of their linkage relationship. The concept of unlinked and completely linked genes. genetic linkage is outlined in this handout and will be discussed in class in the coming weeks. The position of a gene on a chromosome can be Example 1: A dihybrid cross for genes A and B, establish its identity. two unlinked genes. Generation Genotypes Phenotypes Specific Goals P1 AABBXaabb AB and ab 1. Conduct appropriate genetic crosses with a F1 AaBb AB mutant D. melanogaster to determine the following: F2 1AABB:2AABb: 9AB:3Ab:3aB:1ab 2AaBB:4AaBb:1AAbb: 2Aabb:1aaBB:2aaBb:1aabb a. Segregation b. Independent Assortment/Linkage Phenotypic ratio from dihybrid cross for c. -

The Centenary of Janssens's Chiasmatype Theory
PERSPECTIVES The Centenary of Janssens’s Chiasmatype Theory Romain Koszul,*,†,1 Matthew Meselson,‡ Karine Van Doninck,§ Jean Vandenhaute,** and Denise Zickler††,1 *Institut Pasteur, Group Spatial Regulation of Genomes, Department of Genomes and Genetics, F-75015 Paris, France, †2 Centre National de la Recherche Scientifique, Unité Mixte de Recherche (UMR) 3525, F-75015 Paris, France, ‡Department of Molecular and Cellular Biology, Harvard University, Cambridge, MA 02138, §Université de Namur, Facultés universitaires Notre-Dame de la Paix (FUNDP), Unité de Recherche en Biologie des Organismes, B5000 Namur, Belgium, **Université de Namur, FUNDP, Unité de Recherche en Biologie Moléculaire et Département Sciences, Philosophies et Sociétés, B5000 Namur, Belgium, and ††Institut de Génétique et Microbiologie, UMR 8621, Université Paris-Sud, 91405 Orsay, France ABSTRACT The segregation and random assortment of characters observed by Mendel have their basis in the behavior of chromosomes in meiosis. But showing this actually to be the case requires a correct understanding of the meiotic behavior of chromosomes. This was achieved only gradually, over several decades, with much dispute and confusion along the way. One crucial step in the understanding of meiosis was provided in 1909 by Frans Alfons Janssens who published in La Cellule an article entitled “La théorie de la Chiasmatypie. Nouvelle interprétation des cinèses de maturation,” which contains the first description of the chiasma structure. He observed that, of the four chromatids present at the connection sites (chiasmata sites) at diplotene or anaphase of the first meiotic division, two crossed each other and two did not. He therefore postulated that the maternal and paternal chromatids that crossed penetrated the other until they broke and rejoined in maternal and paternal segments new ways; the other two chromatids remained free and thus intact. -

Meiosis in Haploid Rye: Extensive Synapsis and Low Chiasma Frequency
Heredity 73 (1994 580—588 Received 8 February 1994 Genetical Society of Great Britain Meiosis inhaploidrye: extensive synapsis and low chiasma frequency J. L. SANTOS*, M. M. JIMENEZ & M. D1EZ Department of Genetics, Faculty of Biology, Universidad Comp/utense de Madrid, 28040 Madrid, Spain Extensivesynaptonemal complex formation was found at prophase I in whole mount spread preparations of a spontaneous haploid rye, Secale cereale, with values of up to 87.8 per cent of the chromosome complement synapsed. Pairing-partner switches were frequent, giving rise to multiple associations in which all or most of the chromosomes were involved. However, the distribution of synaptonemal complex stretches suggests that synapsis does not occur at random. The frequency of multivalents and the mean frequency of bonded arms at metaphase I were 0.03 and 0.39, respectively. Associations between chromosome arms without heterochromatin were more frequent than between the remaining arms. The observation of recombinant chromosomes for telomeric C-bands at anaphase I indicates that metaphase I bonds are true chiasmata. The correspondence between the location of pairing initiation sites and chiasmata indicates that early synapsis could be confined to homologous regions. Keywords:C-bands,haploid rye, metaphase I bonds, rye, Secale cereale, synapsis. Introduction the case in different monoploids of rye (Levan, 1942; Neijzing, 1982; deJong eta!., 1991). Individualswith chromosome numbers corresponding Pachytene observations by light microscopy to those of the gametes of their species are designated revealed occasional interchromosomal and intra- by the general term 'haploids'. An individual with the chromosomal pairing in monoploids of rice (Chu, gametic chromosome number derived from a diploid 1967), tomato (Ecochard et al., 1969), maize (Ting, species is called 'monoploid' or simply 'haploid', 1966; Ford, 1970; Weber & Alexander, 1972) and whereas the term 'polyhaploid' is used when it is pro- barley (Sadasivaiah & Kasha, 1971, 1973). -
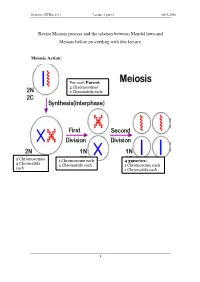
Revise Meiosis Process and the Relation Between Mendel Laws and Meiosis Before Proceeding with This Lecture
Genetics (BTBio 211) Lecture 3 part 2 2015-2016 Revise Meiosis process and the relation between Mendel laws and Meiosis before proceeding with this lecture. Meiosis Action: For each Parent 2 Chromosomes 2 Chromatids each 2 Chromosomes 1 Chromosome each 4 gametes: 4 Chromatid s 4 Chromatids each 1 Chromosome each each 1 Chromatids each 1 Genetics (BTBio 211) Lecture 3 part 2 2015-2016 LINKAGE AND CHROMOSOME MAPPING IN EUKARYOTES I. LINKAGE Genetic linkage is the tendency of genes that are located proximal to each other on a chromosome to be inherited together during meiosis. Genes whose loci are nearer to each other are less likely to be separated onto different chromatids during chromosomal crossover, and are therefore said to be genetically linked. Linked genes: Genes that are inherited together with other gene(s) in form of single unit as they are located on the same chromosome. For example: in fruit flies the genes for eye color and the genes for wing length are on the same chromosome, thus are inherited together. A couple of genes on chromosomes may be present either on the different or on the same chromosome. 1. The independent assortment of two genes located on different chromosomes. Mendel’s Law of Independent Assortment: during gamete formation, segregation of one gene pair is independent of other gene pairs because the traits he studied were determined by genes on different chromosomes. Consider two genes A and B, each with two alleles A a and B b on separate (different) chromosomes. 2 Genetics (BTBio 211) Lecture 3 part 2 2015-2016 Gametes of non-homologous chromosomes assort independently at anaphase producing 4 different genotypes AB, ab, Ab and aB with a genotypic ratio 1:1:1:1. -

Meiosis, Recombination, and Interference
Meiosis, recombination, and interference Karl W Broman Department of Biostatistics Johns Hopkins University Baltimore, Maryland, USA www.biostat.jhsph.edu/˜kbroman Outline Mitosis and meiosis Chiasmata, crossovers Genetic distance Genetic markers, recombination Chromatid and chiasma interference Mather’s formula The count-location model ¢ The gamma model and the ¡ model Data: humans and mice Mitosis: ordinary cell division Chromosomes Chr's pull apart duplicate and cell divides Chromosomes line up Meiosis: production of sex cells Chr's pull apart and cells divide Chr's exchange material and cell divides Chromosomes duplicate Chromosomes pair up The exchange process Vocabulary Four-strand bundle Meiotic products Sister chromatids Non-sister chromatids Chiasma, chiasmata Crossovers Obligate chiasma Genetic distance Two points are d Morgans apart if the average number of crossovers per meiotic product in the intervening interval is d. Usual units: centiMorgan (cM); 100 cM = 1 Morgan ¡ Genetic distance Physical distance The intensity of the crossover process varies by Sex Individual Chromosome Position on chromosome Temperature But we don’t observe crossovers Crossover Crossovers generally not process observeable Marker We instead observe the origin data of DNA at marker loci. odd no. crossovers = recombination event even no. crossovers = no recombination Recombination fraction = Pr(recombination event in interval) Microsatellite markers aka Short Tandem Repeat Polymorphisms (STRPs) GA T AGA T A GA T A CT A TCT A T CT A T Tandem repeat of something like GATA at a specific position in the genome. − Number of repeats varies Use PCR to “amplify” region Use gel electrophoresis to determine length of region + Map functions Connect genetic distance (average no. -

Mycelial Fungi Are Typically Multinucleate (Many Nuclei in a Common
Biology of Fungi Lecture 9: Fungal Genetics, Molecular Genetics, and Genomics Q Mycelial fungi are typically multinucleate (many nuclei in a common cytoplasm) and reap both the benefits of being haploid and a “functional diploid” Q Not so for solely diploid organisms (e.g., Oomycota) and it also different for organisms like S. cerevisiae which can have either a persistent haploid or diploid state u Nonsexual variation: heterokaryosis Q Heterokaryons have two or more genetically different nuclei in a common cytoplasm (as opposed to homokaryons) Q Heterokaryons produced in two ways: l Mutation of a wild-type nucleus l Fusion of two genetically different strains Q Ratio of genetically different nuclei can vary depending upon selection pressures, e.g., Jinks test involving Penicillium cyclopium (see Table 9.3 in Deacon) Q Heterokaryons do break down or segregate into homokaryons by one of two mechanisms l Uninucleate spore production l Branch contains a single nuclear type u Nonsexual variation: parasexuality Q Discovered by Pontecorvo in studying heterokaryosis Q He observed that a heterokaryon having nuclear condition Ab and aB, produced not only parental types (Ab and aB), but also recombinant types, AB and ab. Q Further study of this phenomenon led Pontecorvo to propose a parasexual cycle consisting of three stages: l Diploidization l Mitotic chiasma formation l Haploidization Q Parasexuality in relatively rare and is not a regular cycle as is sexual reproduction u Sexual variation Q Major mechanism for producing recombinants Q Different -

Issue 85 of the Genetics Society Newsletter
JULY 2021 | ISSUE 85 GENETICS SOCIETY NEWS In this issue The Genetics Society News is edited by • Presidential handover Margherita Colucci and items for future • Genetics Society Summer Studentship - Share your story, Part 1 issues can be sent to the editor by email • A chat with Dr Stuart Ritchie - Exploring “Science fictions” to [email protected]. • Committee on Mutagenicity of Chemicals in Food, Consumer The Newsletter is published twice a year, Products and the Environment (COM) with copy dates of July and January. • How genetic linkage was discovered Genetics Society Summer Studentship - Share your stories, Part 1. Read the interviews on page 27 A WORD FROM THE EDITOR A word from the editor Welcome to Issue 85 elcome to the latest issue of the world that cry for change: with WGenetics Society Newsletter! Dr Stuart Ritchie, we explore In this issue, “change” is the keyword. misconduct and fraud in science and the solutions that “open science” Through a series of interviews, we proposes, talking about his latest explored the changes from their book “Science fictions: how fraud, undergraduate role and the career bias, negligence and hype undermine evolution of the past years Summer the search for truth”. Studentship grant winners. Where are they now? What impact that research I would like to draw your attention experience had on them? Find out to the opportunity of contributing more in “Genetics Society Summer to the special issue of Heredity. In Studentship - Share your story, July 2022, this special issue will be Part 1”, page 23. celebrating Mendel’s 200th birthday with short essays, reviews and Big changes happened in the Society research articles on “exceptions” too.