Meiosis, Recombination, and Interference
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Linkage and Genetic Mapping 4TH PAGES © Jones & Bartlett Learning, LLC
© Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION Gene Linkage and © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC 4NOTGenetic FOR SALE OR DISTRIBUTIONMapping NOT FOR SALE OR DISTRIBUTION CHAPTER ORGANIZATION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR4.1 SALELinked OR alleles DISTRIBUTION tend to stay 4.4NOT Polymorphic FOR SALE DNA ORsequences DISTRIBUTION are together in meiosis. 112 used in human genetic mapping. 128 The degree of linkage is measured by the Single-nucleotide polymorphisms (SNPs) frequency of recombination. 113 are abundant in the human genome. 129 The frequency of recombination is the same SNPs in restriction sites yield restriction for coupling and repulsion heterozygotes. 114 fragment length polymorphisms (RFLPs). 130 © Jones & Bartlett Learning,The frequency LLC of recombination differs © Jones & BartlettSimple-sequence Learning, repeats LLC (SSRs) often NOT FOR SALE OR DISTRIBUTIONfrom one gene pair to the next. NOT114 FOR SALEdiffer OR in copyDISTRIBUTION number. 131 Recombination does not occur in Gene dosage can differ owing to copy- Drosophila males. 115 number variation (CNV). 133 4.2 Recombination results from Copy-number variation has helped human populations adapt to a high-starch diet. 134 crossing-over between linked© Jones alleles. & Bartlett Learning,116 LLC 4.5 Tetrads contain© Jonesall & Bartlett Learning, LLC four products of meiosis. -

Integrating Genetic Linkage Maps with Pachytene Chromosome Structure in Maize
Copyright 2004 by the Genetics Society of America Integrating Genetic Linkage Maps With Pachytene Chromosome Structure in Maize Lorinda K. Anderson,*,1 Naser Salameh,† Hank W. Bass,‡ Lisa C. Harper,§ W. Z. Cande,§ Gerd Weber† and Stephen M. Stack* *Department of Biology, Colorado State University, Fort Collins, Colorado 80523, †Department of Plant Breeding and Biotechnology, University of Hohenheim, D-70593 Stuttgart, Germany, ‡Department of Biological Science, Florida State University, Tallahassee, Florida 32306 and §Department of Molecular and Cell Biology, University of California, Berkeley, California 94720 Manuscript received November 4, 2003 Accepted for publication January 9, 2004 ABSTRACT Genetic linkage maps reveal the order of markers based on the frequency of recombination between markers during meiosis. Because the rate of recombination varies along chromosomes, it has been difficult to relate linkage maps to chromosome structure. Here we use cytological maps of crossing over based on recombination nodules (RNs) to predict the physical position of genetic markers on each of the 10 chromosomes of maize. This is possible because (1) all 10 maize chromosomes can be individually identified from spreads of synaptonemal complexes, (2) each RN corresponds to one crossover, and (3) the frequency of RNs on defined chromosomal segments can be converted to centimorgan values. We tested our predic- tions for chromosome 9 using seven genetically mapped, single-copy markers that were independently mapped on pachytene chromosomes using in situ hybridization. The correlation between predicted and observed locations was very strong (r2 ϭ 0.996), indicating a virtual 1:1 correspondence. Thus, this new, high-resolution, cytogenetic map enables one to predict the chromosomal location of any genetically mapped marker in maize with a high degree of accuracy. -

On Parameters of the Human Genome
Journal of Theoretical Biology 288 (2011) 92–104 Contents lists available at ScienceDirect Journal of Theoretical Biology journal homepage: www.elsevier.com/locate/yjtbi On parameters of the human genome Wentian Li The Robert S. Boas Center for Genomics and Human Genetics, The Feinstein Institute for Medical Research, North Shore LIJ Health System, Manhasset, 350 Community Drive, NY 11030, USA article info abstract Article history: There are mathematical constants that describe universal relationship between variables, and physical/ Received 13 April 2011 chemical constants that are invariant measurements of physical quantities. In a similar spirit, we have Received in revised form collected a set of parameters that characterize the human genome. Some parameters have a constant 28 June 2011 value for everybody’s genome, others vary within a limited range. The following nine human genome Accepted 21 July 2011 parameters are discussed here, number of bases (genome size), number of chromosomes (karyotype), Available online 3 August 2011 number of protein-coding gene loci, number of transcription factors, guanine–cytosine (GC) content, Keywords: number of GC-rich gene-rich isochores, density of polymorphic sites, number of newly generated Genome size deleterious mutations in one generation, and number of meiotic crossovers. Comparative genomics and Karyotype theoretical predictions of some parameters are discussed and reviewed. This collection only represents Human genes a beginning of compiling a more comprehensive list of human genome parameters, and knowing these Transcription factors Single nucleotide polymorphisms parameter values is an important part in understanding human evolution. & 2011 Elsevier Ltd. All rights reserved. 1. Introduction never change. Although such assumption has been challenged concerning fundamental physical constants, in particular in the Mathematics, physics, and chemistry all have their standard cosmology context (Dirac, 1937; Gamow, 1967; Varshalovich and sets of basic constants, as expected for any quantitative science. -

Primer on Molecular Genetics
DOE Human Genome Program Primer on Molecular Genetics Date Published: June 1992 U.S. Department of Energy Office of Energy Research Office of Health and Environmental Research Washington, DC 20585 The "Primer on Molecular Genetics" is taken from the June 1992 DOE Human Genome 1991-92 Program Report. The primer is intended to be an introduction to basic principles of molecular genetics pertaining to the genome project. Human Genome Management Information System Oak Ridge National Laboratory 1060 Commerce Park Oak Ridge, TN 37830 Voice: 865/576-6669 Fax: 865/574-9888 E-mail: [email protected] 2 Contents Primer on Molecular Introduction ............................................................................................................. 5 Genetics DNA............................................................................................................................... 6 Genes............................................................................................................................ 7 Revised and expanded Chromosomes ............................................................................................................... 8 by Denise Casey (HGMIS) from the Mapping and Sequencing the Human Genome ...................................... 10 primer contributed by Charles Cantor and Mapping Strategies ..................................................................................................... 11 Sylvia Spengler Genetic Linkage Maps ........................................................................................... -

Genetic Markers, Map Construction, and Their Application in Plant Breeding Jack E
Genetic Markers, Map Construction, and Their Application in Plant Breeding Jack E. Staub1 and Felix C. Serquen2 Vegetable Crops Research, U. S. Department of Agriculture, Agricultural Research Service, Department of Horticulture, University of Wisconsin–Madison, WI 53706 Manju Gupta3 Mycogen Plant Sciences, Madison Laboratories, 5649 East Buckeye Road, Madison, WI 53716 The genetic improvement of a species in a bewildering array of new terms. For scien- RFLPs. Restriction fragment length poly- through artificial selection depends on the tists who have a peripheral interest in genome morphisms (RFLPs) are detected by the use of ability to capitalize on genetic effects that can mapping, but would like to understand the restriction enzymes that cut genomic DNA be distinguished from environmental effects. potential role of MAS in plant improvement, molecules at specific nucleotide sequences Phenotypic selection based on traits that are the wealth of information currently being pro- (restriction sites), thereby yielding variable- conditioned by additive allelic effects can pro- duced in this area can lead to considerable size DNA fragments (Fig. 1). Identification of duce dramatic, economically important confusion. The purpose of this paper is to genomic DNA fragments is made by Southern changes in breeding populations. Genetic describe available marker types and examine blotting, a procedure whereby DNA fragments, markers—heritable entities that are associated factors critical for their use in map construc- separated by electrophoresis, are transferred with economically important traits—can be tion and MAS. This review clarifies how ge- to nitrocellulose or nylon filter (Southern, used by plant breeders as selection tools netic markers are used in map construction 1975). -

The Centenary of Janssens's Chiasmatype Theory
PERSPECTIVES The Centenary of Janssens’s Chiasmatype Theory Romain Koszul,*,†,1 Matthew Meselson,‡ Karine Van Doninck,§ Jean Vandenhaute,** and Denise Zickler††,1 *Institut Pasteur, Group Spatial Regulation of Genomes, Department of Genomes and Genetics, F-75015 Paris, France, †2 Centre National de la Recherche Scientifique, Unité Mixte de Recherche (UMR) 3525, F-75015 Paris, France, ‡Department of Molecular and Cellular Biology, Harvard University, Cambridge, MA 02138, §Université de Namur, Facultés universitaires Notre-Dame de la Paix (FUNDP), Unité de Recherche en Biologie des Organismes, B5000 Namur, Belgium, **Université de Namur, FUNDP, Unité de Recherche en Biologie Moléculaire et Département Sciences, Philosophies et Sociétés, B5000 Namur, Belgium, and ††Institut de Génétique et Microbiologie, UMR 8621, Université Paris-Sud, 91405 Orsay, France ABSTRACT The segregation and random assortment of characters observed by Mendel have their basis in the behavior of chromosomes in meiosis. But showing this actually to be the case requires a correct understanding of the meiotic behavior of chromosomes. This was achieved only gradually, over several decades, with much dispute and confusion along the way. One crucial step in the understanding of meiosis was provided in 1909 by Frans Alfons Janssens who published in La Cellule an article entitled “La théorie de la Chiasmatypie. Nouvelle interprétation des cinèses de maturation,” which contains the first description of the chiasma structure. He observed that, of the four chromatids present at the connection sites (chiasmata sites) at diplotene or anaphase of the first meiotic division, two crossed each other and two did not. He therefore postulated that the maternal and paternal chromatids that crossed penetrated the other until they broke and rejoined in maternal and paternal segments new ways; the other two chromatids remained free and thus intact. -

Meiosis in Haploid Rye: Extensive Synapsis and Low Chiasma Frequency
Heredity 73 (1994 580—588 Received 8 February 1994 Genetical Society of Great Britain Meiosis inhaploidrye: extensive synapsis and low chiasma frequency J. L. SANTOS*, M. M. JIMENEZ & M. D1EZ Department of Genetics, Faculty of Biology, Universidad Comp/utense de Madrid, 28040 Madrid, Spain Extensivesynaptonemal complex formation was found at prophase I in whole mount spread preparations of a spontaneous haploid rye, Secale cereale, with values of up to 87.8 per cent of the chromosome complement synapsed. Pairing-partner switches were frequent, giving rise to multiple associations in which all or most of the chromosomes were involved. However, the distribution of synaptonemal complex stretches suggests that synapsis does not occur at random. The frequency of multivalents and the mean frequency of bonded arms at metaphase I were 0.03 and 0.39, respectively. Associations between chromosome arms without heterochromatin were more frequent than between the remaining arms. The observation of recombinant chromosomes for telomeric C-bands at anaphase I indicates that metaphase I bonds are true chiasmata. The correspondence between the location of pairing initiation sites and chiasmata indicates that early synapsis could be confined to homologous regions. Keywords:C-bands,haploid rye, metaphase I bonds, rye, Secale cereale, synapsis. Introduction the case in different monoploids of rye (Levan, 1942; Neijzing, 1982; deJong eta!., 1991). Individualswith chromosome numbers corresponding Pachytene observations by light microscopy to those of the gametes of their species are designated revealed occasional interchromosomal and intra- by the general term 'haploids'. An individual with the chromosomal pairing in monoploids of rice (Chu, gametic chromosome number derived from a diploid 1967), tomato (Ecochard et al., 1969), maize (Ting, species is called 'monoploid' or simply 'haploid', 1966; Ford, 1970; Weber & Alexander, 1972) and whereas the term 'polyhaploid' is used when it is pro- barley (Sadasivaiah & Kasha, 1971, 1973). -

Uncovering Cryptic Asexuality in Daphnia Magna by RAD Sequencing
GENETICS | INVESTIGATION Uncovering Cryptic Asexuality in Daphnia magna by RAD Sequencing Nils Svendsen,*,1 Celine M. O. Reisser,*,†,1 Marinela Dukic,´ ‡ Virginie Thuillier,† Adeline Ségard,* Cathy Liautard-Haag,† Dominique Fasel,† Evelin Hürlimann,† Thomas Lenormand,* Yan Galimov,§ and Christoph R. Haag*,†,2 *Centre d’Ecologie Fonctionnelle et Evolutive (CEFE)–Unité Mixte de Recherche 5175, Centre National de la Recherche Scientifique (CNRS)–Université de Montpellier–Université Paul-Valéry Montpellier–Ecole Pratique des Hautes Etudes (EPHE), campus CNRS, 19, 34293 Montpellier Cedex 5, France, †Ecology and Evolution, University of Fribourg, 1700 Fribourg, Switzerland, ‡Zoology Institute, Evolutionary Biology, University of Basel, 4051 Basel, Switzerland, and §Koltsov Institute of Developmental Biology, Russian Academy of Sciences, 119334 Moscow, Russia ABSTRACT The breeding systems of many organisms are cryptic and difficult to investigate with observational data, yet they have profound effects on a species’ ecology, evolution, and genome organization. Genomic approaches offer a novel, indirect way to investigate breeding systems, specifically by studying the transmission of genetic information from parents to offspring. Here we exemplify this method through an assessment of self-fertilization vs. automictic parthenogenesis in Daphnia magna. Self-fertilization reduces heterozygosity by 50% compared to the parents, but under automixis, whereby two haploid products from a single meiosis fuse, the expected heterozygosity reduction depends on -
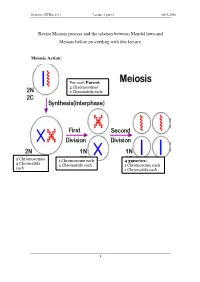
Revise Meiosis Process and the Relation Between Mendel Laws and Meiosis Before Proceeding with This Lecture
Genetics (BTBio 211) Lecture 3 part 2 2015-2016 Revise Meiosis process and the relation between Mendel laws and Meiosis before proceeding with this lecture. Meiosis Action: For each Parent 2 Chromosomes 2 Chromatids each 2 Chromosomes 1 Chromosome each 4 gametes: 4 Chromatid s 4 Chromatids each 1 Chromosome each each 1 Chromatids each 1 Genetics (BTBio 211) Lecture 3 part 2 2015-2016 LINKAGE AND CHROMOSOME MAPPING IN EUKARYOTES I. LINKAGE Genetic linkage is the tendency of genes that are located proximal to each other on a chromosome to be inherited together during meiosis. Genes whose loci are nearer to each other are less likely to be separated onto different chromatids during chromosomal crossover, and are therefore said to be genetically linked. Linked genes: Genes that are inherited together with other gene(s) in form of single unit as they are located on the same chromosome. For example: in fruit flies the genes for eye color and the genes for wing length are on the same chromosome, thus are inherited together. A couple of genes on chromosomes may be present either on the different or on the same chromosome. 1. The independent assortment of two genes located on different chromosomes. Mendel’s Law of Independent Assortment: during gamete formation, segregation of one gene pair is independent of other gene pairs because the traits he studied were determined by genes on different chromosomes. Consider two genes A and B, each with two alleles A a and B b on separate (different) chromosomes. 2 Genetics (BTBio 211) Lecture 3 part 2 2015-2016 Gametes of non-homologous chromosomes assort independently at anaphase producing 4 different genotypes AB, ab, Ab and aB with a genotypic ratio 1:1:1:1. -

Mycelial Fungi Are Typically Multinucleate (Many Nuclei in a Common
Biology of Fungi Lecture 9: Fungal Genetics, Molecular Genetics, and Genomics Q Mycelial fungi are typically multinucleate (many nuclei in a common cytoplasm) and reap both the benefits of being haploid and a “functional diploid” Q Not so for solely diploid organisms (e.g., Oomycota) and it also different for organisms like S. cerevisiae which can have either a persistent haploid or diploid state u Nonsexual variation: heterokaryosis Q Heterokaryons have two or more genetically different nuclei in a common cytoplasm (as opposed to homokaryons) Q Heterokaryons produced in two ways: l Mutation of a wild-type nucleus l Fusion of two genetically different strains Q Ratio of genetically different nuclei can vary depending upon selection pressures, e.g., Jinks test involving Penicillium cyclopium (see Table 9.3 in Deacon) Q Heterokaryons do break down or segregate into homokaryons by one of two mechanisms l Uninucleate spore production l Branch contains a single nuclear type u Nonsexual variation: parasexuality Q Discovered by Pontecorvo in studying heterokaryosis Q He observed that a heterokaryon having nuclear condition Ab and aB, produced not only parental types (Ab and aB), but also recombinant types, AB and ab. Q Further study of this phenomenon led Pontecorvo to propose a parasexual cycle consisting of three stages: l Diploidization l Mitotic chiasma formation l Haploidization Q Parasexuality in relatively rare and is not a regular cycle as is sexual reproduction u Sexual variation Q Major mechanism for producing recombinants Q Different -

Genetic^And Physical Mapping Studies on Mouse Chromosome 2
Genetic^and Physical Mapping Studies on Mouse Chromosome 2 by Stavros Males A thesis submitted for the degree of Doctor of Philosophy at the University of London May 1995 The Galton Laboratory Department of Genetics and Biometry University College London ProQuest Number: 10018553 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. Also, if material had to be removed, a note will indicate the deletion. uest. ProQuest 10018553 Published by ProQuest LLC(2016). Copyright of the Dissertation is held by the Author. All rights reserved. This work is protected against unauthorized copying under Title 17, United States Code. Microform Edition © ProQuest LLC. ProQuest LLC 789 East Eisenhower Parkway P.O. Box 1346 Ann Arbor, Ml 48106-1346 Abstract This thesis describes two genetic maps of mouse chromosome 2 (MMU2), genetic maps relative to the wasted {wsf) and Ragged (Ra) mutations on distal MMU2 and a physical map of the region likely to contain the latter two genes. The first two maps include 18 new PCR markers which were isolated from a subgenomic library constructed from a mouse-hamster somatic cell hybrid line whose main mouse-genome component was MMU2. Fourteen of these loci define microsatellite sequences and four represent randomly chosen DNA sequences. The maps were constructed using two interspecific backcrosses established using a laboratory mouse strain and mice from the related species Mus spretus. The inheritance pattern on MMU2 reveals loci that exhibit significant segregation distortion (SD) in males. -

Conceptualizing Kinship and Dependence on Centimorgan
Volume-03 ISSN: 2455-3085 (Online) Issue-12 RESEARCH REVIEW International Journal of Multidisciplinary December -2018 www.rrjournals.com [UGC Listed Journal] Conceptualizing Kinship and Dependence on Centimorgan *1Archisman Roy and 2Tushar Banerjee *1Student (Author), School of Science, AOSHS, Asansol (India) 2Research Scholar, University, Department of Zoology, SSSUTMS, Bhopal (India) ARTICLE DETAILS ABSTRACT Article History Genetics has been one of the most interesting and wildly applauded subject areas among Published Online: 10 December 2018 biologists. Precisely this subject deal with genes and the prime purpose of this biological element is to create new sanguine relations. Our paper encounters an intriguing way of Keywords analyzing kinship and its origin in genetics. This piece of research firstly goes through Genes, Chromosomes, loci, Morgan, introducing the whole context in detail with the right citations and refereeing validated modeling, homologous. researches, followingly it pacifies the sources of data with enough lucidity. The main text contains conceptualizing the whole subject matter quite engagingly as well as ponders over *Corresponding Author a detailed description of how kinship depends on centimorgans and loci of different alleles. Email:archismanroy2002[at]gmail.com Next to that overall summary of the previous elucidations have been assorted to precede a precise and qualitative analysis of the whole report. At last, the write-up ends with the complete texture of lucidity and contents relating to sanguine kinship and its dependence on the position of genes with centimorgans. 1. Introduction genes already existing thereon chromosome, the It is obvious to have so many confusions within the minds method is understood as “recombination”. of the many enthusiastic genealogists on precisely what a III.