Table S-1. Identification of Species, Lineage, and Gene ID Used for Multiple Sequence Alignment All Sequences Are from Genomes Unless
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Phototrophic Oxidation of Ferrous Iron by a Rhodomicrobium Vannielii Strain
Phototrophic oxidation of ferrous iron by a Rhodomicrobium vannielii strain Silke Heising and Bernhard Schink Author for correspondence: Bernhard Schink. Tel: 49 7531 882140. Fax: 49 7531 882966. e-mail: Bernhard.Schink!uni-konstanz.de Fakulta$ tfu$r Biologie, Oxidation of ferrous iron was studied with the anaerobic phototrophic Universita$ t Konstanz, bacterial strain BS-1. Based on morphology, substrate utilization patterns, Postfach 5560, D-78434 Konstanz, Germany arrangement of intracytoplasmic membranes and the in vivo absorption spectrum, this strain was assigned to the known species Rhodomicrobium vannielii. Also, the type strain of this species oxidized ferrous iron in the light. Phototrophic growth of strain BS-1 with ferrous iron as electron donor was stimulated by the presence of acetate or succinate as cosubstrates. The ferric iron hydroxides produced precipitated on the cell surfaces as solid crusts which impeded further iron oxidation after two to three generations. The complexing agent nitrilotriacetate stimulated iron oxidation but the yield of cell mass did not increase stoichiometrically under these conditions. Other complexing agents inhibited cell growth. Ferric iron was not reduced in the dark, and manganese salts were neither oxidized nor reduced. It is concluded that ferrous iron oxidation by strain BS-1 is only a side activity of this bacterium that cannot support growth exclusively with this electron source over prolonged periods of time. Keywords: iron metabolism, phototrophic bacteria, Rhodomicrobium vannielii, iron complexation, nitrilotriacetate (NTA) INTRODUCTION anoxygenic purple bacteria, including a Rhodo- microbium vannielii-like isolate (Widdel et al., 1993). Iron is the fourth most important element in the Earth’s Other strains of purple phototrophs able to oxidize crust, making up about 5% of the total crust mass ferrous iron are strain L7, a non-motile rod with gas (Ehrlich, 1990). -

A Study on the Phototrophic Microbial Mat Communities of Sulphur Mountain Thermal Springs and Their Association with the Endangered, Endemic Snail Physella Johnsoni
A Study on the Phototrophic Microbial Mat Communities of Sulphur Mountain Thermal Springs and their Association with the Endangered, Endemic Snail Physella johnsoni By Michael Bilyj A thesis submitted to the Faculty of Graduate Studies in partial fulfillment of the requirements for the degree of Master of Science Department of Microbiology Faculty of Science University of Manitoba Winnipeg, Manitoba October 2011 © Copyright 2011, Michael A. Bilyj 1 Abstract The seasonal population fluctuation of anoxygenic phototrophs and the diversity of cyanobacteria at the Sulphur Mountain thermal springs of Banff, Canada were investigated and compared to the drastic population changes of the endangered snail Physella johnsoni. A new species and two strains of Rhodomicrobium were taxonomically characterized in addition to new species of Rhodobacter and Erythromicrobium. Major mat-forming organisms included Thiothrix-like species, oxygenic phototrophs of genera Spirulina, Oscillatoria, and Phormidium and purple nonsulfur bacteria Rhodobacter, Rhodopseudomonas and Rhodomicrobium. Aerobic anoxygenic phototrophs comprised upwards of 9.6 x 104 CFU/cm2 of mat or 18.9% of total aerobic heterotrophic bacterial isolates at certain sites, while maximal purple nonsulfur and purple sulfur bacteria were quantified at 3.2 x 105 and 2.0 x 106 CFU/cm2 of mat, respectively. Photosynthetic activity measurements revealed incredibly productive carbon fixation rates averaging 40.5 mg C/cm2/24 h. A temporal mismatch was observed for mat area and prokaryote-based organics to P. johnsoni population flux in a ―tracking inertia‖ manner. 2 Acknowledgements It is difficult to express sufficient gratitude to my supervisor Dr. Vladimir Yurkov for his unfaltering patience, generosity and motivation throughout this entire degree. -

Diversity of Understudied Archaeal and Bacterial Populations of Yellowstone National Park: from Genes to Genomes Daniel Colman
University of New Mexico UNM Digital Repository Biology ETDs Electronic Theses and Dissertations 7-1-2015 Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes Daniel Colman Follow this and additional works at: https://digitalrepository.unm.edu/biol_etds Recommended Citation Colman, Daniel. "Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes." (2015). https://digitalrepository.unm.edu/biol_etds/18 This Dissertation is brought to you for free and open access by the Electronic Theses and Dissertations at UNM Digital Repository. It has been accepted for inclusion in Biology ETDs by an authorized administrator of UNM Digital Repository. For more information, please contact [email protected]. Daniel Robert Colman Candidate Biology Department This dissertation is approved, and it is acceptable in quality and form for publication: Approved by the Dissertation Committee: Cristina Takacs-Vesbach , Chairperson Robert Sinsabaugh Laura Crossey Diana Northup i Diversity of understudied archaeal and bacterial populations from Yellowstone National Park: from genes to genomes by Daniel Robert Colman B.S. Biology, University of New Mexico, 2009 DISSERTATION Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biology The University of New Mexico Albuquerque, New Mexico July 2015 ii DEDICATION I would like to dedicate this dissertation to my late grandfather, Kenneth Leo Colman, associate professor of Animal Science in the Wool laboratory at Montana State University, who even very near the end of his earthly tenure, thought it pertinent to quiz my knowledge of oxidized nitrogen compounds. He was a man of great curiosity about the natural world, and to whom I owe an acknowledgement for his legacy of intellectual (and actual) wanderlust. -

Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline
Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline • Phlogenetics of Archaea • Phlogenetics of archaeal lipids • Papers Phyla • Two? main phyla – Euryarchaeota • Methanogens • Extreme halophiles • Extreme thermophiles • Sulfate-reducing – Crenarchaeota • Extreme thermophiles – Korarchaeota? • Hyperthermophiles • indicated only by environmental DNA sequences – Nanoarchaeum? • N. equitans a fast evolving euryarchaeal lineage, not novel, early diverging archaeal phylum – Ancient archael group? • In deepest brances of Crenarchaea? Euryarchaea? Archaeal Lipids • Methanogens – Di- and tetra-ethers of glycerol and isoprenoid alcohols – Core mostly archaeol or caldarchaeol – Core sometimes sn-2- or Images removed due to sn-3-hydroxyarchaeol or copyright considerations. macrocyclic archaeol –PMI • Halophiles – Similar to methanogens – Exclusively synthesize bacterioruberin • Marine Crenarchaea Depositional Archaeal Lipids Biological Origin Environment Crocetane methanotrophs? methane seeps? methanogens, PMI (2,6,10,15,19-pentamethylicosane) methanotrophs hypersaline, anoxic Squalane hypersaline? C31-C40 head-to-head isoprenoids Smit & Mushegian • “Lost” enzymes of MVA pathway must exist – Phosphomevalonate kinase (PMK) – Diphosphomevalonate decarboxylase – Isopentenyl diphosphate isomerase (IPPI) Kaneda et al. 2001 Rohdich et al. 2001 Boucher et al. • Isoprenoid biosynthesis of archaea evolved through a combination of processes – Co-option of ancestral enzymes – Modification of enzymatic specificity – Orthologous and non-orthologous gene -

Archaeology of Eukaryotic DNA Replication
Downloaded from http://cshperspectives.cshlp.org/ on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press Archaeology of Eukaryotic DNA Replication Kira S. Makarova and Eugene V. Koonin National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Maryland 20894 Correspondence: [email protected] Recent advances in the characterization of the archaeal DNA replication system together with comparative genomic analysis have led to the identification of several previously un- characterized archaeal proteins involved in replication and currently reveal a nearly com- plete correspondence between the components of the archaeal and eukaryotic replication machineries. It can be inferred that the archaeal ancestor of eukaryotes and even the last common ancestor of all extant archaea possessed replication machineries that were compa- rable in complexity to the eukaryotic replication system. The eukaryotic replication system encompasses multiple paralogs of ancestral components such that heteromeric complexes in eukaryotes replace archaeal homomeric complexes, apparently along with subfunctionali- zation of the eukaryotic complex subunits. In the archaea, parallel, lineage-specific dupli- cations of many genes encoding replication machinery components are detectable as well; most of these archaeal paralogs remain to be functionally characterized. The archaeal rep- lication system shows remarkable plasticity whereby even some essential components such as DNA polymerase and single-stranded DNA-binding protein are displaced by unrelated proteins with analogous activities in some lineages. ouble-stranded DNA is the molecule that Okazaki fragments (Kornberg and Baker 2005; Dcarries genetic information in all cellular Barry and Bell 2006; Hamdan and Richardson life-forms; thus, replication of this genetic ma- 2009; Hamdan and van Oijen 2010). -
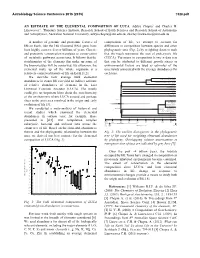
An Estimate of the Elemental Composition of Luca
Astrobiology Science Conference 2015 (2015) 7328.pdf AN ESTIMATE OF THE ELEMENTAL COMPOSITION OF LUCA. Aditya Chopra1 and Charles H. Lineweaver1, 1Planetary Science Institute, Research School of Earth Sciences and Research School of Astronomy and Astrophysics, Australian National University, [email protected], [email protected] A number of genomic and proteomic features of composition of life, we attempt to account for life on Earth, like the 16S ribosomal RNA gene, have differences in composition between species and other been highly conserved over billions of years. Genetic phylogenetic taxa (Fig. 2) by weighting datasets such and proteomic conservation translates to conservation that the result represents the root of prokaryotic life of metabolic pathways across taxa. It follows that the (LUCA). Variations in composition between data sets stoichiometry of the elements that make up some of that can be attributed to different growth stages or the biomolecules will be conserved. By extension, the environmental factors are used as estimates of the elemental make up of the whole organism is a uncertainty associated with the average abundances for relatively conserved feature of life on Earth [1,2]. each taxa. We describe how average bulk elemental Euryarchaeota 1a Methanococci, Methanobacteria, Methanopyri 7 Euryarchaeota 1b abundances in extant life can yield an indirect estimate 6 Thermoplasmata, Methanomicrobia, Halobacteria, Archaeoglobi Euryarchaeota 2 4 Thermococci of relative abundances of elements in the Last Crenarchaeota Sulfolobus, Thermoproteus ? Thaumarchaeota Universal Common Ancestor (LUCA). The results Cenarchaeum ? Korarchaeota ARMAN could give us important hints about the stoichiometry ? 2 Archaeal Richmond Mine Acidophilic Nanoorganisms Nanoarchaeota of the environment where LUCA existed and perhaps Archaea Terrabacteria clues to the processes involved in the origin and early Actinobacteria, Deinococcus-Thermus, 1 Cyanobacteria, Life Chloroflexi, 9 evolution of life [3]. -

Universidad Politécnica De Madrid Escuela Técnica Superior De Ingeniería Agronómica Alimentaria Y De Biosistemas Departamento De Biotecnología-Biología Vegetal
UNIVERSIDAD POLITÉCNICA DE MADRID ESCUELA TÉCNICA SUPERIOR DE INGENIERÍA AGRONÓMICA ALIMENTARIA Y DE BIOSISTEMAS DEPARTAMENTO DE BIOTECNOLOGÍA-BIOLOGÍA VEGETAL TESIS DOCTORAL Caracterización de una nueva proteína hipertermófila NifB y su papel en el mecanismo de síntesis de NifB-co, precursor del grupo metálico de FeMo-co Author: Alessandro Scandurra Director: Prof. Luis Manuel Rubio Herrero Co-director: Dr. Simon Jean Marius Arragain i ii UNIVERSIDAD POLITÉCNICA DE MADRID ESCUELA TÉCNICA SUPERIOR DE INGENIERÍA AGRONÓMICA ALIMENTARIA Y DE BIOSISTEMAS DEPARTAMENTO DE BIOTECNOLOGÍA-BIOLOGÍA VEGETAL Memoria presentada por D. Alessandro Scandurra para optar al grado de Doctor Director Dr. Luis Manuel Rubio Herrero Profesor Titular UPM Madrid, 2017 © This copy oF the thesis has been supplied on condition that anyone who consults it is understood to recognize that its copyright rest with the author and that no quotation From the thesis, or any inFormation derived therefrom may be published without the author’s prior, written consent. I II AcKnowledgments As each journey, also this one has an end (Thanks goodness). It was a long trip, with several changes oF direction. Like For many other people, the PhD adventure was not easy, but I have to admit that I was lucky enough to meet really unique people, that became special Friendships. First oF all, I want to thank my Family, especially my mother and my Father, who always push me to go Forward in my life and to Follow my dreams, ready to pay the sacrifice oF the distance. This expirience has helped me to understand how easy is to be a son and how challenging is to be a parent. -

Pan-Genome Analysis and Ancestral State Reconstruction Of
www.nature.com/scientificreports OPEN Pan‑genome analysis and ancestral state reconstruction of class halobacteria: probability of a new super‑order Sonam Gaba1,2, Abha Kumari2, Marnix Medema 3 & Rajeev Kaushik1* Halobacteria, a class of Euryarchaeota are extremely halophilic archaea that can adapt to a wide range of salt concentration generally from 10% NaCl to saturated salt concentration of 32% NaCl. It consists of the orders: Halobacteriales, Haloferaciales and Natriabales. Pan‑genome analysis of class Halobacteria was done to explore the core (300) and variable components (Softcore: 998, Cloud:36531, Shell:11784). The core component revealed genes of replication, transcription, translation and repair, whereas the variable component had a major portion of environmental information processing. The pan‑gene matrix was mapped onto the core‑gene tree to fnd the ancestral (44.8%) and derived genes (55.1%) of the Last Common Ancestor of Halobacteria. A High percentage of derived genes along with presence of transformation and conjugation genes indicate the occurrence of horizontal gene transfer during the evolution of Halobacteria. A Core and pan‑gene tree were also constructed to infer a phylogeny which implicated on the new super‑order comprising of Natrialbales and Halobacteriales. Halobacteria1,2 is a class of phylum Euryarchaeota3 consisting of extremely halophilic archaea found till date and contains three orders namely Halobacteriales4,5 Haloferacales5 and Natrialbales5. Tese microorganisms are able to dwell at wide range of salt concentration generally from 10% NaCl to saturated salt concentration of 32% NaCl6. Halobacteria, as the name suggests were once considered a part of a domain "Bacteria" but with the discovery of the third domain "Archaea" by Carl Woese et al.7, it became part of Archaea. -
![Downloaded from the NCBI FTP Site [37]](https://docslib.b-cdn.net/cover/4020/downloaded-from-the-ncbi-ftp-site-37-1134020.webp)
Downloaded from the NCBI FTP Site [37]
Life 2015, 5, 818-840; doi:10.3390/life5010818 OPEN ACCESS life ISSN 2075-1729 www.mdpi.com/journal/life Article Archaeal Clusters of Orthologous Genes (arCOGs): An Update and Application for Analysis of Shared Features between Thermococcales, Methanococcales, and Methanobacteriales Kira S. Makarova *, Yuri I. Wolf and Eugene V. Koonin National Center for Biotechnology Information, NLM, National Institutes of Health, Bethesda, MD 20894, USA; E-Mails: [email protected] (Y.I.W.); [email protected] (E.V.K.) * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +1-301-435-5913; Fax: +1-301-435-7793. Academic Editors: Hans-Peter Klenk, Michael W. W. Adams and Roger A. Garrett Received: 12 January 2015 / Accepted: 28 February 2015 / Published: 10 March 2015 Abstract: With the continuously accelerating genome sequencing from diverse groups of archaea and bacteria, accurate identification of gene orthology and availability of readily expandable clusters of orthologous genes are essential for the functional annotation of new genomes. We report an update of the collection of archaeal Clusters of Orthologous Genes (arCOGs) to cover, on average, 91% of the protein-coding genes in 168 archaeal genomes. The new arCOGs were constructed using refined algorithms for orthology identification combined with extensive manual curation, including incorporation of the results of several completed and ongoing research projects in archaeal genomics. A new level of classification is introduced, superclusters that unit two or more arCOGs and more completely reflect gene family evolution than individual, disconnected arCOGs. Assessment of the current archaeal genome annotation in public databases indicates that consistent use of arCOGs can significantly improve the annotation quality. -

Deoxyribonucleic Acid Base Sequence Homologies of Some Budding and Prosthecate Bacterla RICHARD L
JOURNAL OF BACTERIOLOGY, Apr. 1972, p. 256-261 Vol. 110, No. 1 Copyright © 1972 American Society for Microbiology Printed in U.S.A. Deoxyribonucleic Acid Base Sequence Homologies of Some Budding and Prosthecate Bacterla RICHARD L. MOORE' AND PETER HIRSCH2 Department of Microbiology and Public Health, Michigan State University, East Lansing, Michigan 48823 Received for publication 21 December 1971 The genetic relatedness of a number of budding and prosthecate bacteria was determined by deoxyribonucleic acid (DNA) homology experiments of the di- rect binding type. Strains of Hyphomicrobium sp. isolated from aquatic habi- tats were found to have relatedness values ranging from 9 to 70% with strain "EA-617," a subculture of the Hyphomicrobium isolated by Mevius from river water. Strains obtained from soil enrichments had lower values with EA-617, ranging from 3 to 5%. Very little or no homology was detected between the amino acid-utilizing strain Hyphomicrobium neptunium and other Hyphomi- crobium strains, although significant homology was observed with the two Hyphomonas strains examined. No homology could be detected between pros- thecate bacteria of the genera Rhodomicrobium, Prosthecomicrobium, Ancal- omicrobium, or Caulobacter, and Hyphomicrobium strain EA-617 or H. nep- tunium LE-670. The grouping of Hyphomicrobium strains by their relatedness values agrees well with a grouping according to the base composition of their DNA species. It is concluded that bacteria possessing cellular extensions repre- sent a widely diverse group of organisms. Two genera of bacteria, Hyphomicrobium drum, P. pneumaticum, Ancalomicrobium adetum, and Rhodomicrobium, are listed under the and Caulobacter crescentus were obtained from J. T. family of Hyphomicrobiaceae in the seventh Staley (Seattle); Rhodomicrobium vannielii was re- edition of Bergey's Manual of Determinative ceived from H. -

WARREN LYNWOOD GARDNER Expression Vectors for the Methane-Producing Archaeon Methanococcus Maripaludis (Under the Direction of WILLIAM B
WARREN LYNWOOD GARDNER Expression vectors for the methane-producing archaeon Methanococcus maripaludis (Under the Direction of WILLIAM B. WHITMAN) Methanogens are strict anaerobes that use one and two carbon compounds for methanogenesis. They contain many unusual cofactors and enzymes, and many of their proteins are oxygen-sensitive and are present at low concentrations within the cells. An expression system that overexpresses homologous and heterologous proteins would facilitate research on the unique oxygen-sensitive enzymes in these organisms. However, traditional expression systems may lack the cofactors and maturation enzymes necessary for the expression of the active holoenzymes. Therefore, a major goal of this work was to develop expression vectors. To develop a shuttle vector, a series of integrative vectors were first constructed. The integrative vectors were based on the pUC-derivative pMEB.2. pMEB.2 provided the puromycin resistance marker for methanococci, an Escherichia coli origin of replication, and an ampicillin resistance marker for E. coli. A multiple cloning site and the Methanococcus voltae histone promoter (PhmvA) were added to form the integrative, expression vector pWLG14. To demonstrate the utility of PhmvA, pWLG14 was used to overexpress the genomic copy of the Methanococcus maripaludis acetohydroxyacid synthase. To form an expression shuttle vector suitable for heterologous genes, pWLG14 and the cryptic plasmid pURB500 from M. maripaludis strain were ligated together to form pWLG30. pWLG30 was the first expression shuttle vector for the methanogens. To demonstrate the utility of pWLG30, the E. coli b-galactosidase gene was cloned into pWLG30 to yield pWLG30+lacZ. Upon transformation into M. maripaludis, the recombinant strain expressed b-galactosidase to the level of 1% of the cellular protein. -

Research Collection
Research Collection Doctoral Thesis Development and application of molecular tools to investigate microbial alkaline phosphatase genes in soil Author(s): Ragot, Sabine A. Publication Date: 2016 Permanent Link: https://doi.org/10.3929/ethz-a-010630685 Rights / License: In Copyright - Non-Commercial Use Permitted This page was generated automatically upon download from the ETH Zurich Research Collection. For more information please consult the Terms of use. ETH Library DISS. ETH NO.23284 DEVELOPMENT AND APPLICATION OF MOLECULAR TOOLS TO INVESTIGATE MICROBIAL ALKALINE PHOSPHATASE GENES IN SOIL A thesis submitted to attain the degree of DOCTOR OF SCIENCES of ETH ZURICH (Dr. sc. ETH Zurich) presented by SABINE ANNE RAGOT Master of Science UZH in Biology born on 25.02.1987 citizen of Fribourg, FR accepted on the recommendation of Prof. Dr. Emmanuel Frossard, examiner PD Dr. Else Katrin Bünemann-König, co-examiner Prof. Dr. Michael Kertesz, co-examiner Dr. Claude Plassard, co-examiner 2016 Sabine Anne Ragot: Development and application of molecular tools to investigate microbial alkaline phosphatase genes in soil, c 2016 ⃝ ABSTRACT Phosphatase enzymes play an important role in soil phosphorus cycling by hydrolyzing organic phosphorus to orthophosphate, which can be taken up by plants and microorgan- isms. PhoD and PhoX alkaline phosphatases and AcpA acid phosphatase are produced by microorganisms in response to phosphorus limitation in the environment. In this thesis, the current knowledge of the prevalence of phoD and phoX in the environment and of their taxonomic distribution was assessed, and new molecular tools were developed to target the phoD and phoX alkaline phosphatase genes in soil microorganisms.