Intronic Mirna Mir-3666 Modulates Its Host Gene FOXP2 Functions In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Small Cell Ovarian Carcinoma: Genomic Stability and Responsiveness to Therapeutics
Gamwell et al. Orphanet Journal of Rare Diseases 2013, 8:33 http://www.ojrd.com/content/8/1/33 RESEARCH Open Access Small cell ovarian carcinoma: genomic stability and responsiveness to therapeutics Lisa F Gamwell1,2, Karen Gambaro3, Maria Merziotis2, Colleen Crane2, Suzanna L Arcand4, Valerie Bourada1,2, Christopher Davis2, Jeremy A Squire6, David G Huntsman7,8, Patricia N Tonin3,4,5 and Barbara C Vanderhyden1,2* Abstract Background: The biology of small cell ovarian carcinoma of the hypercalcemic type (SCCOHT), which is a rare and aggressive form of ovarian cancer, is poorly understood. Tumourigenicity, in vitro growth characteristics, genetic and genomic anomalies, and sensitivity to standard and novel chemotherapeutic treatments were investigated in the unique SCCOHT cell line, BIN-67, to provide further insight in the biology of this rare type of ovarian cancer. Method: The tumourigenic potential of BIN-67 cells was determined and the tumours formed in a xenograft model was compared to human SCCOHT. DNA sequencing, spectral karyotyping and high density SNP array analysis was performed. The sensitivity of the BIN-67 cells to standard chemotherapeutic agents and to vesicular stomatitis virus (VSV) and the JX-594 vaccinia virus was tested. Results: BIN-67 cells were capable of forming spheroids in hanging drop cultures. When xenografted into immunodeficient mice, BIN-67 cells developed into tumours that reflected the hypercalcemia and histology of human SCCOHT, notably intense expression of WT-1 and vimentin, and lack of expression of inhibin. Somatic mutations in TP53 and the most common activating mutations in KRAS and BRAF were not found in BIN-67 cells by DNA sequencing. -

Case Report a Fetus with Kabuki Syndrome 2 Detected by Chromosomal Microarray Analysis
Int J Clin Exp Pathol 2020;13(2):302-306 www.ijcep.com /ISSN:1936-2625/IJCEP0104334 Case Report A fetus with Kabuki syndrome 2 detected by chromosomal microarray analysis Chen-Zhao Lin1, Bi-Ru Qi1, Jian-Su Hu2, Xiu-Qiong Huang3 Departments of 1Obstetrics and Gynecology, 2Ultrasound, 3Laboratory Medicine, Fuzhou Municipal First Hospital Affiliated to Fujian Medical University, Fuzhou 350009, Fujian Province, The People’s Republic of China Received November 1, 2019; Accepted January 26, 2020; Epub February 1, 2020; Published February 15, 2020 Abstract: Background: Kabuki syndrome is a rare multiple congenital anomaly syndrome characterized by distinct facial features, intellectual disability, cardiovascular and musculoskeletal abnormalities, persistence of fetal finger- tip pads, and postnatal growth deficiency. Currently, the diagnosis mainly depends on clinical manifestations and genetic testing. To date, there is no report on the identification Kabuki syndrome in fetuses using chromosomal microarray analysis (CMA). Case presentation: A fetus was identified with growth retardation and cardiovascular abnormality on color Doppler ultrasonography; however, non-invasive prenatal testing (NIPT) revealed a low risk and G-banding karyotyping revealed no abnormal karyotype detected. CMA identified a 1.3 Mb deletion on the X chromosome (Xp11.3) containing KDM6A, DUSP21, MIR222, MIR221 and CXorf36 genes. The fetus was diagnosed with Kabuki syndrome 2, and labor was induced. In addition, CMA detected a 1.3 Mb deletion in the chromosome Xp11.3 in the mother, which contains 5 genes namely KDM6A, DUSP21, MIR222, MIR221 and CXorf36, while no chromosomal abnormality was identified in the father. Conclusions: We report a fetus with Kabuki syndrome 2 detected using CMA. -
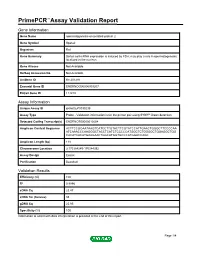
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name spermatogenesis-associated protein 2 Gene Symbol Spata2 Organism Rat Gene Summary Sertoli cell mRNA expression is induced by FSH; may play a role in spermatogenesis; localized to the nucleus Gene Aliases Not Available RefSeq Accession No. Not Available UniGene ID Rn.201291 Ensembl Gene ID ENSRNOG00000009207 Entrez Gene ID 114210 Assay Information Unique Assay ID qRnoCEP0030238 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENSRNOT00000012604 Amplicon Context Sequence ACTTCCGGAATAAGTCATCCTTGTACTTCGTATCCATTGAACTGGGCTTCCCCAA ATCAAACCCAAGGGCTACCTCATCTCCCCCATGGCTCTGGGGCTGGAGGCTGG CACATCACATGAAGAACTGGCATGGTGCCCATGGACCAGC Amplicon Length (bp) 118 Chromosome Location 3:170384245-170384392 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 100 R2 0.9996 cDNA Cq 23.47 cDNA Tm (Celsius) 85 gDNA Cq 25.95 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report Spata2, Rat Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

(HPG) Axis Published: 18 April 2017 Matthew D
www.nature.com/scientificreports OPEN Widespread patterns of sexually dimorphic gene expression in an avian hypothalamic–pituitary– Received: 30 November 2016 Accepted: 16 February 2017 gonadal (HPG) axis Published: 18 April 2017 Matthew D. MacManes1, Suzanne H. Austin2, Andrew S. Lang1, April Booth2, Victoria Farrar2 & Rebecca M. Calisi2 The hypothalamic-pituitary-gonadal (HPG) axis is a key biological system required for reproduction and associated sexual behaviors to occur. In the avian reproductive model of the rock dove (Columba livia), we characterized the transcript community of each tissue of the HPG axis in both sexes, thereby significantly expanding our mechanistic insight into HPG activity. We report greater sex-biased differential expression in the pituitary as compared to the hypothalamus, with multiple genes more highly expressed in the male pituitary being related to secretory function, and multiple genes more highly expressed in the female pituitary being related to reproduction, growth, and development. We report tissue-specific and sex-biased expression in genes commonly investigated when studying reproduction, highlighting the need for sex parity in future studies. In addition, we uncover new targets of investigation in both sexes, which could potentially change our understanding of HPG function. The hypothalamic-pituitary-gonadal (HPG) axis is a system comprised of endocrine glands whose function is vital to the regulation of reproduction and associated behaviors (Fig. 1). In all vertebrates studied, from humans to Agnatha, the jawless fishes, the HPG axis is present and its function is generally conserved1. For reproduc- tion to occur, the hypothalamus must produce and secrete gonadotropin-releasing hormone (GnRH), which causes the pituitary gland to secrete gonadotropins, luteinizing hormone (LH) and follicle stimulating hormone (FSH)2. -

Itcs) in the Mouse Amygdala of Tshz1 Mutants Correlates with Fear, Depression, and Social Interaction Phenotypes
1160 • The Journal of Neuroscience, January 31, 2018 • 38(5):1160–1177 Development/Plasticity/Repair Loss of Intercalated Cells (ITCs) in the Mouse Amygdala of Tshz1 Mutants Correlates with Fear, Depression, and Social Interaction Phenotypes X Jeffrey Kuerbitz,1 Melinda Arnett,5 Sarah Ehrman,1 XMichael T. Williams,3 XCharles V. Vorhees,3 X Simon E. Fisher,6,7 Alistair N. Garratt,8 XLouis J. Muglia,5 Ronald R. Waclaw,1,4 and XKenneth Campbell1,2 Divisions of 1Developmental Biology, 2Neurosurgery, 3Neurology, 4Experimental Hematology and Cancer Biology, 5Center for Prevention of Preterm Birth, Perinatal Institute, Cincinnati Children’s Hospital Medical Center, University of Cincinnati College of Medicine, Cincinnati, OH 45229, 6Language and Genetics Department, Max Planck Institute for Psycholinguistics, 6500 AH Nijmegen, The Netherlands, 7Donders Institute for Brain, Cognition and Behaviour, Radboud University, Nijmegen, The Netherlands, and 8Institute of Cell Biology and Neurobiology, Center for Anatomy, Charite´ University Hospital Berlin, 10117 Berlin, Germany The intercalated cells (ITCs) of the amygdala have been shown to be critical regulatory components of amygdalar circuits, which control appropriate fear responses. Despite this, the molecular processes guiding ITC development remain poorly understood. Here we establish the zinc finger transcription factor Tshz1 as a marker of ITCs during their migration from the dorsal lateral ganglionic eminence through maturity. Using germline and conditional knock-out (cKO) mouse models, we show that Tshz1 is required for the proper migration and differentiation of ITCs. In the absence of Tshz1, migrating ITC precursors fail to settle in their stereotypical locations encapsulating the lateral amygdala and BLA. Furthermore, they display reductions in the ITC marker Foxp2 and ectopic persistence of the dorsal lateral ganglionic eminence marker Sp8. -

Mediator of DNA Damage Checkpoint 1 (MDC1) Is a Novel Estrogen Receptor Co-Regulator in Invasive 6 Lobular Carcinoma of the Breast 7 8 Evelyn K
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.16.423142; this version posted December 16, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 1 Running Title: MDC1 co-regulates ER in ILC 2 3 Research article 4 5 Mediator of DNA damage checkpoint 1 (MDC1) is a novel estrogen receptor co-regulator in invasive 6 lobular carcinoma of the breast 7 8 Evelyn K. Bordeaux1+, Joseph L. Sottnik1+, Sanjana Mehrotra1, Sarah E. Ferrara2, Andrew E. Goodspeed2,3, James 9 C. Costello2,3, Matthew J. Sikora1 10 11 +EKB and JLS contributed equally to this project. 12 13 Affiliations 14 1Dept. of Pathology, University of Colorado Anschutz Medical Campus 15 2Biostatistics and Bioinformatics Shared Resource, University of Colorado Comprehensive Cancer Center 16 3Dept. of Pharmacology, University of Colorado Anschutz Medical Campus 17 18 Corresponding author 19 Matthew J. Sikora, PhD.; Mail Stop 8104, Research Complex 1 South, Room 5117, 12801 E. 17th Ave.; Aurora, 20 CO 80045. Tel: (303)724-4301; Fax: (303)724-3712; email: [email protected]. Twitter: 21 @mjsikora 22 23 Authors' contributions 24 MJS conceived of the project. MJS, EKB, and JLS designed and performed experiments. JLS developed models 25 for the project. EKB, JLS, SM, and AEG contributed to data analysis and interpretation. SEF, AEG, and JCC 26 developed and performed informatics analyses. MJS wrote the draft manuscript; all authors read and revised the 27 manuscript and have read and approved of this version of the manuscript. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Genome-Wide Identification and Analysis of Prognostic Features in Human Cancers
bioRxiv preprint doi: https://doi.org/10.1101/2021.06.01.446243; this version posted June 1, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Genome-wide identification and analysis of prognostic features in human cancers Joan C. Smith1,2 and Jason M. Sheltzer1* 1. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 11724. 2. Google, Inc., New York, NY 10011. * Lead contact; to whom correspondence may be addressed. E-mail: [email protected]. bioRxiv preprint doi: https://doi.org/10.1101/2021.06.01.446243; this version posted June 1, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Abstract Clinical decisions in cancer rely on precisely assessing patient risk. To improve our ability to accurately identify the most aggressive malignancies, we constructed genome-wide survival models using gene expression, copy number, methylation, and mutation data from 10,884 patients with known clinical outcomes. We identified more than 100,000 significant prognostic biomarkers and demonstrate that these genomic features can predict patient outcomes in clinically-ambiguous situations. While adverse biomarkers are commonly believed to represent cancer driver genes and promising therapeutic targets, we show that cancer features associated with shorter survival times are not enriched for either oncogenes or for successful drug targets. -

1 Supporting Information for a Microrna Network Regulates
Supporting Information for A microRNA Network Regulates Expression and Biosynthesis of CFTR and CFTR-ΔF508 Shyam Ramachandrana,b, Philip H. Karpc, Peng Jiangc, Lynda S. Ostedgaardc, Amy E. Walza, John T. Fishere, Shaf Keshavjeeh, Kim A. Lennoxi, Ashley M. Jacobii, Scott D. Rosei, Mark A. Behlkei, Michael J. Welshb,c,d,g, Yi Xingb,c,f, Paul B. McCray Jr.a,b,c Author Affiliations: Department of Pediatricsa, Interdisciplinary Program in Geneticsb, Departments of Internal Medicinec, Molecular Physiology and Biophysicsd, Anatomy and Cell Biologye, Biomedical Engineeringf, Howard Hughes Medical Instituteg, Carver College of Medicine, University of Iowa, Iowa City, IA-52242 Division of Thoracic Surgeryh, Toronto General Hospital, University Health Network, University of Toronto, Toronto, Canada-M5G 2C4 Integrated DNA Technologiesi, Coralville, IA-52241 To whom correspondence should be addressed: Email: [email protected] (M.J.W.); yi- [email protected] (Y.X.); Email: [email protected] (P.B.M.) This PDF file includes: Materials and Methods References Fig. S1. miR-138 regulates SIN3A in a dose-dependent and site-specific manner. Fig. S2. miR-138 regulates endogenous SIN3A protein expression. Fig. S3. miR-138 regulates endogenous CFTR protein expression in Calu-3 cells. Fig. S4. miR-138 regulates endogenous CFTR protein expression in primary human airway epithelia. Fig. S5. miR-138 regulates CFTR expression in HeLa cells. Fig. S6. miR-138 regulates CFTR expression in HEK293T cells. Fig. S7. HeLa cells exhibit CFTR channel activity. Fig. S8. miR-138 improves CFTR processing. Fig. S9. miR-138 improves CFTR-ΔF508 processing. Fig. S10. SIN3A inhibition yields partial rescue of Cl- transport in CF epithelia. -

The Role of Microrna in Human Leukemia: a Review
Leukemia (2009) 23, 1257–1263 & 2009 Macmillan Publishers Limited All rights reserved 0887-6924/09 $32.00 www.nature.com/leu SPOTLIGHT REVIEW The role of microRNA in human leukemia: a review S Yendamuri1 and GA Calin2 1Department of Thoracic Surgery, Roswell Park Cancer Institute, Buffalo, NY, USA and 2Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, TX, USA MicroRNAs (miRNAs or miRs) are 18–22-nucleotide non-coding target. If the sequence is a perfect match, the final result seems RNAs that have emerged as a new paradigm of epigenetic to be target mRNA degradation in a manner similar to siRNA- regulation in both normal development and cellular function, and in the pathogenesis of human disease including cancer. induced gene expression silencing. This seems to be the This review summarizes the current literature of mechanism of predominant mechanism in plants. A less than perfect match gene regulation by miRNA and their role in hematopoiesis and may result in inefficient translation leading to effective down- leukemogenesis. An understanding of these processes sug- regulation of the gene, a mechanism more common in gests further avenues for research to understand gene regula- animals.7–9 Isolated examples of miRNA leading to upregulation tion and miRNA-based therapeutic approaches. of genes have been reported.10 Leukemia (2009) 23, 1257–1263; doi:10.1038/leu.2008.382; The mechanism of translational repression is known only published online 15 January 2009 11 Keywords: microRNA; non-coding RNA; microarray; partially. The mRNA segments that miRNAs bind seem to be mostly in the 30UTRs of genes. -

Anti-EIF2C1 / AGO1 Antibody (ARG63916)
Product datasheet [email protected] ARG63916 Package: 100 μg anti-EIF2C1 / AGO1 antibody Store at: -20°C Summary Product Description Goat Polyclonal antibody recognizes EIF2C1 / AGO1 Tested Reactivity Hu Predict Reactivity Ms, Rat, Dog Tested Application IHC-P, WB Specificity This product is not expected to cross-react with EIF2C2, EIF2C3 and EIF2C4. Host Goat Clonality Polyclonal Isotype IgG Target Name EIF2C1 / AGO1 Antigen Species Human Immunogen C-KNASYNLDPYIQEF Conjugation Un-conjugated Alternate Names GERP95; Q99; eIF2C 1; EIF2C; EIF2C1; Argonaute RISC catalytic component 1; Argonaute1; Protein argonaute-1; hAgo1; Putative RNA-binding protein Q99; eIF-2C 1; Eukaryotic translation initiation factor 2C 1 Application Instructions Application table Application Dilution IHC-P 2.5 µg/ml WB 0.3 - 1 µg/ml Application Note WB: Recommend incubate at RT for 1h. IHC-P: Antigen Retrieval: Steam tissue section in Citrate buffer (pH 6.0). * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Calculated Mw 97 kDa Properties Form Liquid Purification Purified from goat serum by ammonium sulphate precipitation followed by antigen affinity chromatography using the immunizing peptide. Buffer Tris saline (pH 7.3), 0.02% Sodium azide and 0.5% BSA Preservative 0.02% Sodium azide Stabilizer 0.5% BSA www.arigobio.com 1/3 Concentration 0.5 mg/ml Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C or below. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. -

Targeted Pharmacological Therapy Restores Β-Cell Function for Diabetes Remission
Targeted pharmacological therapy restores -cell function for diabetes remission Sachs, Stephan; Bastidas-Ponce, Aimée; Tritschler, Sophie; Bakhti, Mostafa; Böttcher, Anika; Sánchez-Garrido, Miguel A; Tarquis-Medina, Marta; Kleinert, Maximilian; Fischer, Katrin; Jall, Sigrid; Harger, Alexandra; Bader, Erik; Roscioni, Sara; Ussar, Siegfried; Feuchtinger, Annette; Yesildag, Burcak; Neelakandhan, Aparna; Jensen, Christine B; Cornu, Marion; Yang, Bin; Finan, Brian; DiMarchi, Richard D; Tschöp, Matthias H; Theis, Fabian J; Hofmann, Susanna M.; Müller, Timo D; Lickert, Heiko Published in: Nature Metabolism DOI: 10.1038/s42255-020-0171-3 Publication date: 2020 Document version Publisher's PDF, also known as Version of record Document license: CC BY Citation for published version (APA): Sachs, S., Bastidas-Ponce, A., Tritschler, S., Bakhti, M., Böttcher, A., Sánchez-Garrido, M. A., Tarquis-Medina, M., Kleinert, M., Fischer, K., Jall, S., Harger, A., Bader, E., Roscioni, S., Ussar, S., Feuchtinger, A., Yesildag, B., Neelakandhan, A., Jensen, C. B., Cornu, M., ... Lickert, H. (2020). Targeted pharmacological therapy restores - cell function for diabetes remission. Nature Metabolism, 2(2), 192-209. https://doi.org/10.1038/s42255-020- 0171-3 Download date: 05. Oct. 2021 ARTICLES https://doi.org/10.1038/s42255-020-0171-3 There are amendments to this paper Targeted pharmacological therapy restores β-cell function for diabetes remission Stephan Sachs1,2,3,4,19, Aimée Bastidas-Ponce1,4,5,6,19, Sophie Tritschler1,4,7,8,19, Mostafa Bakhti 1,4,5, Anika Böttcher1,4,5, Miguel A. Sánchez-Garrido2, Marta Tarquis-Medina1,4,5,6, Maximilian Kleinert2,9, Katrin Fischer2,3, Sigrid Jall2,3, Alexandra Harger2, Erik Bader1, Sara Roscioni1, Siegfried Ussar 4,6,10, Annette Feuchtinger11, Burcak Yesildag12, Aparna Neelakandhan12, Christine B.