Rejoinder for Discussion on “Horseshoe-Based Bayesian Nonparametric Estimation of Effective Population Size Trajectories”
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Horseshoe-Based Bayesian Nonparametric Estimation of Effective Population Size Trajectories
Received: 13 August 2018 Accepted: 9 July 2019 DOI: 10.1111/biom.13276 BIOMETRIC METHODOLOGY Horseshoe-based Bayesian nonparametric estimation of effective population size trajectories James R. Faulkner1,2 Andrew F. Magee3 Beth Shapiro4,5 Vladimir N. Minin6 1Quantitative Ecology and Resource Abstract Management, University of Washington, Seattle, Washington Phylodynamics is an area of population genetics that uses genetic sequence data to 2Fish Ecology Division, Northwest Fisheries estimate past population dynamics. Modern state-of-the-art Bayesian nonparamet- Science Center, National Marine Fisheries ric methods for recovering population size trajectories of unknown form use either Service, NOAA, Seattle, Washington change-point models or Gaussian process priors. Change-point models suffer from 3Department of Biology, University of Washington, Seattle, Washington computational issues when the number of change-points is unknown and needs to 4Ecology and Evolutionary Biology be estimated. Gaussian process-based methods lack local adaptivity and cannot accu- Department and Genomics Institute, rately recover trajectories that exhibit features such as abrupt changes in trend or vary- University of California Santa Cruz, Santa Cruz, California ing levels of smoothness. We propose a novel, locally adaptive approach to Bayesian 5Howard Hughes Medical Institute, nonparametric phylodynamic inference that has the flexibility to accommodate a University of California Santa Cruz, large class of functional behaviors. Local adaptivity results from modeling the log- Santa Cruz, California transformed effective population size a priori as a horseshoe Markov random field, 6Department of Statistics, University of arecentlyproposedstatisticalmodelthatblendstogetherthebestpropertiesofthe California Irvine, Irvine, California change-point and Gaussian process modeling paradigms. We use simulated data to Correspondence assess model performance, and find that our proposed method results in reduced bias Vladimir N. -

Division Or Research Center Department Faculty Description
Division or Research Center Include in 2019 Department Faculty Description Sust. Research Reason for excluding (Y/N/M) Anderson's current research incorporates computer technologies to engage questions Y about land use and social interventions into the environment. His recent work, Silicon Monuments - in collaboration with the Silicon Valley Toxics Coalition - uses augmented reality software on hand-held devices to create a site-specific, multimedia documentary about toxic Superfund sites in Silicon Valley. Viewers can explore the sites and interact with the documentary, which reveals hidden environmental damage and its health and social costs. Website link: http://arts.ucsc.edu/faculty/eanderson/ Arts Art Elliott W. Anderson A. Laurie Palmer’s work is concerned with material explorations of matter’s active Y nature as it asserts itself on different scales and in different speeds, and with collaborating on strategic actions in the contexts of social and environmental justice. These two directions sometimes run parallel and sometimes converge, taking form as sculpture, installation, writing, and public projects. Collaboration, with other humans and with non-humans, is a central ethic in her practice. Website link: http: //alauriepalmer.net/ Arts Art Laurie Palmer Contemporary art and visual culture, investigating in particular the diverse ways that Y artists and activists have negotiated crises associated with globalization, including the emerging conjunction of post-9/11 political sovereignty and statelessness, the hauntings of the colonial past, and the growing biopolitical conflicts around ecology and climate change. Most recently Demos is the author of Decolonizing Nature: Contemporary Art and the Politics of Ecology (Sternberg Press, 2016), which investigates how concern for ecological crisis has entered the field of contemporary art and visual culture in recent years, and considers art and visual cultural practices globally. -

SFU Thesis Template Files
Reconstructing Northern Fur Seal Population Diversity through Ancient and Modern DNA Data by Cara Leanne Halseth B.Sc., University of Northern British Columbia, 2011 Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of Master of Arts in the Department of Archaeology Faculty of Environment © Cara Leanne Halseth 2015 SIMON FRASER UNIVERSITY Summer 2015 Approval Name: Cara Leanne Halseth Degree: Master of Arts Title: Reconstructing Northern Fur Seal Population Diversity through Ancient and Modern DNA Data Examining Committee: Chair: Catherine D‟Andrea Professor Dongya Yang Senior Supervisor Professor Deborah C. Merrett Supervisor Adjunct Professor Iain McKechnie External Examiner SSHRC Postdoctoral Fellowship Dept of Anthropology University of Oregon Date Defended/Approved: July 2, 2015 ii Abstract Archaeological and historic evidence suggests that northern fur seal (Callorhinus ursinus) has undergone several population and distribution changes (including commercial sealing) potentially resulting in a loss of genetic diversity and population structure. This study analyzes 36 unpublished mtDNA sequences from archaeological sites 1900-150 BP along the Pacific Northwest Coast from Moss et al. (2006) as well as published data (primarily Pinsky et al. [2010]) to investigate this species‟ genetic diversity and population genetics in the past. The D-loop data shows high nucleotide and haplotype diversity, with continuity of two separate subdivisions (haplogroups) through time. Nucleotide mismatch analysis suggests population expansion in both ancient and modern data. AMOVA analysis (FST and ΦST) reveals some „structure‟ detectable between several archaeological sites. While the data reviewed here did not reveal dramatic patterning, the AMOVA analysis does identify several significant FST values, indicating some level of ancient population „structure‟, which deserves future study. -

Reunion and Symposium the Past, Present, and Future of Ecology at Uga
SCHEDULE AT A GLANCE Jan. 12-14, 2018 REUNION AND SYMPOSIUM THE PAST, PRESENT, AND FUTURE OF ECOLOGY AT UGA FRIDAY, JAN. 12, 2018 4:00 – 7:00 p.m. “First” Friday Welcoming Reception (Ecology Building/Ecology Tent) The reunion symposium officially begins with a welcome reception featuring food, beverages, music and the opportunity to catch up with friends and classmates. (OK, technically it’s second Friday.) Registration and packet pick-up will be available in the Ecology lobby. Odum School Art Exhibit (Ecology Seminar Room) 5:30 – 6:00 p.m. Ecotones “UGA's ecologically-minded co-ed a cappella group” (Ecology Auditorium) 6:00 p.m. Slideshow of alumni and current & past faculty (Ecology Auditorium) SATURDAY, JAN. 13, 2018 7:30 – 8:30 a.m. Breakfast & Registration (Forestry Auditorium) Coffee and pastries available 8:30 – 10:00 a.m. Session I: Opening Plenary Session (Forestry Auditorium) • Welcome from DEAN JOHN GITTLEMAN • PAMELA WHITTEN | Senior Vice President for Academic Affairs and Provost • PETER RAVEN | President Emeritus of the Missouri Botanical Garden and former Home Secretary of the National Academy of Sciences Ecosystems and the Ecology of Change Plenary Address by Monica Turner, PhD ‘85 MONICA TURNER, a member of the National Academy of Sciences since 2004 and Past President of the Ecological Society of America, is a landscape ecologist who studies the ecosystem effects of fire and other disturbances and the ecological effects of climate and land use change. 10:00 – 10:15 a.m. Coffee Break (Ecology Tent) 10:15 – 11:30 a.m. Session II: Foundational Research and Current Connections: Alumni Perspectives (Forestry) • CHRISTOPHER D’ELIA, PhD ‘74 • WEIXIN CHENG, PhD ‘89 • EVELYN GAISER, PhD ‘97 • CHRISTINA FAUST, B.S./M.S., ‘09 11:30 – 12:00 Ecology and the University of Georgia Jere Morehead, President, University of Georgia p.m. -
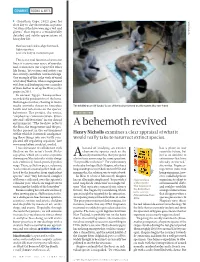
A Behemoth Revived
COMMENT BOOKS & ARTS (Jonathan Cape, 2012) goes for close day-to-day observation, in poems “written at the hive wearing a veil and gloves” that express a wonderfully detailed and subtle appreciation of RIA NOVOSTI/SPL honeybee life: How bees touch and re-align their touch. Light migration; noise of a body in continual repair This is one vital function of art in our lives: it restores our sense of wonder, and so increases our respect for other life forms. Yet writers and artists can also actively contribute new knowledge. One example of this is the work of visual artist Amy Shelton, whose engagement with bees and beekeeping over a number of years led her to set up the Honeyscribe project in 2011. In ancient Egypt, ‘honeyscribes’ recorded the productivity of the hives. Shelton goes further, charting in multi media artworks threats to honeybee The 40,000-year-old ‘Lyuba’ is one of the best-preserved woolly mammoths ever found. health and reflections on the species’ behaviour. Her project, she writes, DE-EXTINCTION “emphasizes communication, diver sity and collaboration” in our shared environment. “The beehive reflects the flora, the temperature and the pes A behemoth revived ticides present in the environment within which it is situated, amalgamat Henry Nicholls examines a clear appraisal of what it ing these things into one vastly com would really take to resurrect extinct species. plicated self-regulating organism” (see www.amyshelton.co.uk/art_works). I was fortunate to collaborate with hazard of studying an extinct has a place in our Shelton on the artist’s book Melis- charismatic species such as the scientific future, but sographia, which sets a series of poems woolly mammoth is that you spend not as an antidote to drawing on Maeterlinck’s study along Aa lot of time answering the same question: extinctions that have side embossed, hand-painted pollen “Is it possible to clone it?” For evolutionary already occurred,” maps. -

Career Pathway Tracker 35 Years of Supporting Early Career Research Fellows Contents
Career pathway tracker 35 years of supporting early career research fellows Contents President’s foreword 4 Introduction 6 Scientific achievements 8 Career achievements 14 Leadership 20 Commercialisation 24 Public engagement 28 Policy contribution 32 How have the fellowships supported our alumni? 36 Who have we supported? 40 Where are they now? 44 Research Fellowship to Fellow 48 Cover image: Graphene © Vertigo3d CAREER PATHWAY TRACKER 3 President’s foreword The Royal Society exists to encourage the development and use of Very strong themes emerge from the survey About this report science for the benefit of humanity. One of the main ways we do that about why alumni felt they benefited. The freedom they had to pursue the research they This report is based on the first is by investing in outstanding scientists, people who are pushing the wanted to do because of the independence Career Pathway Tracker of the alumni of University Research Fellowships boundaries of our understanding of ourselves and the world around the schemes afford is foremost in the minds of respondents. The stability of funding and and Dorothy Hodgkin Fellowships. This us and applying that understanding to improve lives. flexibility are also highly valued. study was commissioned by the Royal Society in 2017 and delivered by the Above Thirty-five years ago, the Royal Society The vast majority of alumni who responded The Royal Society has long believed in the Careers Research & Advisory Centre Venki Ramakrishnan, (CRAC), supported by the Institute for President of the introduced our University Research Fellowships to the survey – 95% of University Research importance of identifying and nurturing the Royal Society. -

Bayesian Phylogenetic and Phylodynamic Data Integration Using BEAST 1.10 Marc A
Virus Evolution, 2018, 4(1): vey016 doi: 10.1093/ve/vey016 Resources Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10 Marc A. Suchard,1,2,3,*,† Philippe Lemey,4,‡ Guy Baele,4,§ Daniel L. Ayres,5 Alexei J. Drummond,6,7,* and Andrew Rambaut8,*,** 1Department of Biomathematics, David Geffen School of Medicine, University of California, Los Angeles, 621 Charles E. Young Dr., South, Los Angeles, CA, 90095 USA, 2Department of Biostatistics, Fielding School of Public Health, University of California, Los Angeles, 650 Charles E, Young Dr., South, Los Angeles, CA, 90095 USA, 3Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, 695 Charles E. Young Dr., South, Los Angeles, CA, 90095 USA, 4Department of Microbiology and Immunology, Rega Institute, KU Leuven, Herestraat 49, 3000 Leuven, Belgium, 5Center for Bioinformatics and Computational Biology, University of Maryland, College Park, 125 Biomolecular Science Bldg #296, College Park, MD 20742 USA, , 6Department of Computer Science, University of Auckland, 303/38 Princes St., Auckland, 1010 NZ, 7Centre for Computational Evolution, University of Auckland, 303/38 Princes St., Auckland, 1010 NZ and 8Institute of Evolutionary Biology, University of Edinburgh, Ashworth Laboratories, Edinburgh, EH9 3FL UK *Corresponding author: E-mail: [email protected] (M.A.S.); [email protected] (A.J.D.); [email protected] (A.R.) †http://orcid.org/0000-0001-9818-479X ‡http://orcid.org/0000-0003-2826-5353 §http://orcid.org/0000-0002-1915-7732 **http://orcid.org/0000-0003-4337-3707 Abstract The Bayesian Evolutionary Analysis by Sampling Trees (BEAST) software package has become a primary tool for Bayesian phylogenetic and phylodynamic inference from genetic sequence data. -

A New Genus of Horse from Pleistocene North America
RESEARCH ARTICLE A new genus of horse from Pleistocene North America Peter D Heintzman1,2*, Grant D Zazula3, Ross DE MacPhee4, Eric Scott5,6, James A Cahill1, Brianna K McHorse7, Joshua D Kapp1, Mathias Stiller1,8, Matthew J Wooller9,10, Ludovic Orlando11,12, John Southon13, Duane G Froese14, Beth Shapiro1,15* 1Department of Ecology and Evolutionary Biology, University of California, Santa Cruz, Santa Cruz, United States; 2Tromsø University Museum, UiT - The Arctic University of Norway, Tromsø, Norway; 3Yukon Palaeontology Program, Government of Yukon, Whitehorse, Canada; 4Department of Mammalogy, Division of Vertebrate Zoology, American Museum of Natural History, New York, United States; 5Cogstone Resource Management, Incorporated, Riverside, United States; 6California State University San Bernardino, San Bernardino, United States; 7Department of Organismal and Evolutionary Biology, Harvard University, Cambridge, United States; 8Department of Translational Skin Cancer Research, German Consortium for Translational Cancer Research, Essen, Germany; 9College of Fisheries and Ocean Sciences, University of Alaska Fairbanks, Fairbanks, United States; 10Alaska Stable Isotope Facility, Water and Environmental Research Center, University of Alaska Fairbanks, Fairbanks, United States; 11Centre for GeoGenetics, Natural History Museum of Denmark, København K, Denmark; 12Universite´ Paul Sabatier, Universite´ de Toulouse, Toulouse, France; 13Keck-CCAMS Group, Earth System Science Department, University of California, Irvine, Irvine, United States; 14Department of Earth and Atmospheric Sciences, University of Alberta, Edmonton, Canada; 15UCSC Genomics Institute, University of California, Santa Cruz, Santa Cruz, United States *For correspondence: [email protected] (PDH); [email protected] (BS) Competing interests: The Abstract The extinct ‘New World stilt-legged’, or NWSL, equids constitute a perplexing group authors declare that no of Pleistocene horses endemic to North America. -

M Ethods in M Olecular B Iology
M ETHODS IN M OLECULAR B IOLOGY Series Editor John M. Walker School of Life and Medical Sciences University of Hertfordshire Hatfield, Hertfordshire, AL10 9AB, UK For further volumes: http://www.springer.com/series/7651 Ancient DNA Methods and Protocols Second Edition Edited by Beth Shapiro Department of Ecology and Evolutionary Biology, University of California Santa Cruz, Santa Cruz, CA, USA Axel Barlow Institute for Biochemistry and Biology, University of Potsdam, Potsdam, Germany Peter D. Heintzman Tromsø University Museum, UiT—The Arctic University of Norway, Tromsø, Norway Michael Hofreiter, Johanna L. A. Paijmans Institute for Biochemistry and Biology, University of Potsdam, Potsdam, Germany André E. R. Soares Laboratório Nacional de Computação Científica, Petrópolis, RJ, Brazil Editors Beth Shapiro Axel Barlow Department of Ecology and Evolutionary Institute for Biochemistry and Biology Biology University of Potsdam University of California Santa Cruz Potsdam, Germany Santa Cruz, CA, USA Michael Hofreiter Peter D. Heintzman Institute for Biochemistry and Biology Tromsø University Museum University of Potsdam UiT—The Arctic University of Norway Potsdam, Germany Tromsø, Norway Andre´ E. R. Soares Johanna L. A. Paijmans Laborato´rio Nacional de Computac¸a˜o Cientı´fica Institute for Biochemistry and Biology Petro´polis, RJ, Brazil University of Potsdam Potsdam, Germany ISSN 1064-3745 ISSN 1940-6029 (electronic) Methods in Molecular Biology ISBN 978-1-4939-9175-4 ISBN 978-1-4939-9176-1 (eBook) https://doi.org/10.1007/978-1-4939-9176-1 -

Demographic Reconstruction from Ancient DNA Supports Rapid
RESEARCH ARTICLE Demographic reconstruction from ancient DNA supports rapid extinction of the great auk Jessica E Thomas1,2‡*, Gary R Carvalho1†, James Haile2, Nicolas J Rawlence3, Michael D Martin4, Simon YW Ho5, Arno´ r Þ Sigfu´sson6, Vigfu´s A Jo´ sefsson6, Morten Frederiksen7, Jannie F Linnebjerg7, Jose A Samaniego Castruita2, Jonas Niemann2, Mikkel-Holger S Sinding2,8, Marcela Sandoval-Velasco2, Andre´ ER Soares9, Robert Lacy10, Christina Barilaro11, Juila Best12,13, Dirk Brandis14, Chiara Cavallo15, Mikelo Elorza16, Kimball L Garrett17, Maaike Groot18, Friederike Johansson19, Jan T Lifjeld20, Go¨ ran Nilson19, Dale Serjeanston21, Paul Sweet22, Errol Fuller23, Anne Karin Hufthammer24, Morten Meldgaard25, Jon Fjeldsa˚ 26, Beth Shapiro9, Michael Hofreiter27, John R Stewart28†, M Thomas P Gilbert2,4†, Michael Knapp29†* 1Molecular Ecology and Fisheries Genetics Laboratory, School of Biological Sciences, Bangor University, Bangor, United Kingdom; 2Natural History Museum of Denmark, University of Copenhagen, Copenhagen, Denmark; 3Otago Palaeogenetics Laboratory, Department of Zoology, University of Otago, Dunedin, New Zealand; 4Department of Natural History, University Museum, Norwegian University of Science and Technology, Trondheim, Norway; 5School of Life and 6 *For correspondence: Environmental Sciences, University of Sydney, Sydney, Australia; Verkı´s Consulting 7 [email protected] (JET); Engineers, Reykjavik, Iceland; Department of Bioscience, Aarhus University, [email protected] (MK) Roskilde, Denmark; 8Greenland Institute -

DOI: 10.1126/Science.1101074 , 1561 (2004); 306 Science , Et Al. Beth
Rise and Fall of the Beringian Steppe Bison Beth Shapiro, et al. Science 306, 1561 (2004); DOI: 10.1126/science.1101074 This copy is for your personal, non-commercial use only. If you wish to distribute this article to others, you can order high-quality copies for your colleagues, clients, or customers by clicking here. Permission to republish or repurpose articles or portions of articles can be obtained by following the guidelines here. The following resources related to this article are available online at www.sciencemag.org (this infomation is current as of April 25, 2011 ): Updated information and services, including high-resolution figures, can be found in the online version of this article at: http://www.sciencemag.org/content/306/5701/1561.full.html Supporting Online Material can be found at: http://www.sciencemag.org/content/suppl/2004/11/22/306.5701.1561.DC1.html on April 25, 2011 A list of selected additional articles on the Science Web sites related to this article can be found at: http://www.sciencemag.org/content/306/5701/1561.full.html#related This article has been cited by 154 article(s) on the ISI Web of Science This article has been cited by 50 articles hosted by HighWire Press; see: http://www.sciencemag.org/content/306/5701/1561.full.html#related-urls This article appears in the following subject collections: www.sciencemag.org Ecology http://www.sciencemag.org/cgi/collection/ecology Downloaded from Science (print ISSN 0036-8075; online ISSN 1095-9203) is published weekly, except the last week in December, by the American Association for the Advancement of Science, 1200 New York Avenue NW, Washington, DC 20005. -

Beth Shapiro Curriculum Vitae
Beth Shapiro Curriculum Vitae Department of Ecology and Evolutionary Biology Born: 14 Jan 1976 University of California Santa Cruz Citizenship: USA Office: 144 Biomedical Sciences Mailstop: EE Biology / EMS Santa Cruz, CA 95064 USA Tel: +1 (831) 459-3003 [email protected] http://pgl.soe.ucsc.edu Education: DPhil (Zoology) Oxford University, 2003. MS (Ecology) University of Georgia, 1999. BS (Ecology) University of Georgia, 1999. Current Position: Associate Professor, Ecology and Evolutionary Biology, UC Santa Cruz, (2012- present) Past Positions: Shaffer Associate Professor, Department of Biology, The Pennsylvania State University (2011-2012) Shaffer Assistant Professor, Department of Biology, The Pennsylvania State University (2007- 2011) Director, Henry Wellcome Ancient Biomolecules Centre, Oxford University (2004-2007) Royal Society University Research Fellow, Oxford University (2006-2007) Wellcome Trust Research Fellow, Oxford University (2004-2006) Selected Honors and Awards: Packard Fellow, 2010 PopTech Science and Public Leadership Fellow, 2010 National Geographic Emerging Explorer, 2010 MacArthur Fellow, 2009 Searle Scholar, 2009 Visiting Fellow, Isaac Newton Institute for Mathematical Sciences, University of Cambridge, 2007 Smithsonian Young Innovator, 2007 University Research Fellow, The Royal Society, 2006 Research Fellow, Balliol College, University of Oxford, 2002-2007 Rhodes Scholar, 1999 Scientific Service: Associate Editor, Molecular Biology and Evolution (2007- current). Guest Editor: Proceedings of the National Academy of Sciences USA. Scientific Advisory Board: Museum of Beringia, Dawson City, YT. Organizer: Genome10K 2015. Member: Public Interfaces of the Life Sciences Roundtable, The National Academies (2013-2015) Panelist: Public Interfaces of Science, The National Academies. Panelist: Molecular Evolution and Genomics funding panel, Genes and Genome Systems Cluster, Division of Molecular and Cellular Biosciences, The National Science Foundation.