(Panthera Leo Ssp.) Reveals Three Distinct Taxa and a Late Pleistocene Reduction in Genetic Diversity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Horseshoe-Based Bayesian Nonparametric Estimation of Effective Population Size Trajectories
Received: 13 August 2018 Accepted: 9 July 2019 DOI: 10.1111/biom.13276 BIOMETRIC METHODOLOGY Horseshoe-based Bayesian nonparametric estimation of effective population size trajectories James R. Faulkner1,2 Andrew F. Magee3 Beth Shapiro4,5 Vladimir N. Minin6 1Quantitative Ecology and Resource Abstract Management, University of Washington, Seattle, Washington Phylodynamics is an area of population genetics that uses genetic sequence data to 2Fish Ecology Division, Northwest Fisheries estimate past population dynamics. Modern state-of-the-art Bayesian nonparamet- Science Center, National Marine Fisheries ric methods for recovering population size trajectories of unknown form use either Service, NOAA, Seattle, Washington change-point models or Gaussian process priors. Change-point models suffer from 3Department of Biology, University of Washington, Seattle, Washington computational issues when the number of change-points is unknown and needs to 4Ecology and Evolutionary Biology be estimated. Gaussian process-based methods lack local adaptivity and cannot accu- Department and Genomics Institute, rately recover trajectories that exhibit features such as abrupt changes in trend or vary- University of California Santa Cruz, Santa Cruz, California ing levels of smoothness. We propose a novel, locally adaptive approach to Bayesian 5Howard Hughes Medical Institute, nonparametric phylodynamic inference that has the flexibility to accommodate a University of California Santa Cruz, large class of functional behaviors. Local adaptivity results from modeling the log- Santa Cruz, California transformed effective population size a priori as a horseshoe Markov random field, 6Department of Statistics, University of arecentlyproposedstatisticalmodelthatblendstogetherthebestpropertiesofthe California Irvine, Irvine, California change-point and Gaussian process modeling paradigms. We use simulated data to Correspondence assess model performance, and find that our proposed method results in reduced bias Vladimir N. -

Mizzoualumnus1975novp16-19.Pdf (3.318Mb)
THE GAUE THAT TRAPPED HISTORY Natural Trap Cave has been collecting bones of unwary animals for at least 13,000 years. Expeditions led by a Mizzou anthropologist are digging up those bones and clues about the cycles of climatic change. Text and Photos by Dave Holman IIi / m ISSOllRI ilLUrnrus The only entrenee to Natural Trap Ceve la the hole In the roof. Any enlmal that might have survived the fallatlll became a victim. Workera at theahe, below, ehherrappellnto theeave or descend theacafloldlng. A herd of small horses stampeded through the tall grass across a plateau near the edge of a canyon, pur sued by a large, long-legged cat. The cat closed on the slowest horse, forcing her along the canyon edge onto a peninsula of limestone. The cat sprang, fas tened its claws and teeth in the horse's neck, and suddenly, horse and cat disappeared from the face of the earth. Twelve thousand years later, a green panel truck and a dusty jeep bounced along a dirt road over the same plateau, now covered with sage and prickly pear. On the limestone peninsula where the horse and cat disappeared, the vehicles stopped by a 15- foot-wide hole in the rock. Eighty feet below is the floor of Natural Trap Cave. The cave floor is covered with the accumulated dust of centuries, clearly stratified and containing thou sands of bones of animals that failed to see the hole. This summer was the second consecutive year that Bob Gilbert, research associate in Mizzou's an thropology department had led an organized ex pedition to the trap. -

Division Or Research Center Department Faculty Description
Division or Research Center Include in 2019 Department Faculty Description Sust. Research Reason for excluding (Y/N/M) Anderson's current research incorporates computer technologies to engage questions Y about land use and social interventions into the environment. His recent work, Silicon Monuments - in collaboration with the Silicon Valley Toxics Coalition - uses augmented reality software on hand-held devices to create a site-specific, multimedia documentary about toxic Superfund sites in Silicon Valley. Viewers can explore the sites and interact with the documentary, which reveals hidden environmental damage and its health and social costs. Website link: http://arts.ucsc.edu/faculty/eanderson/ Arts Art Elliott W. Anderson A. Laurie Palmer’s work is concerned with material explorations of matter’s active Y nature as it asserts itself on different scales and in different speeds, and with collaborating on strategic actions in the contexts of social and environmental justice. These two directions sometimes run parallel and sometimes converge, taking form as sculpture, installation, writing, and public projects. Collaboration, with other humans and with non-humans, is a central ethic in her practice. Website link: http: //alauriepalmer.net/ Arts Art Laurie Palmer Contemporary art and visual culture, investigating in particular the diverse ways that Y artists and activists have negotiated crises associated with globalization, including the emerging conjunction of post-9/11 political sovereignty and statelessness, the hauntings of the colonial past, and the growing biopolitical conflicts around ecology and climate change. Most recently Demos is the author of Decolonizing Nature: Contemporary Art and the Politics of Ecology (Sternberg Press, 2016), which investigates how concern for ecological crisis has entered the field of contemporary art and visual culture in recent years, and considers art and visual cultural practices globally. -

A Middle Holocene Steppe Bison and Paleoenvironments from the Versleuce Meadows, Whitehorse, Yukon, Canada Grant D
1138 ARTICLE A middle Holocene steppe bison and paleoenvironments from the Versleuce Meadows, Whitehorse, Yukon, Canada Grant D. Zazula, Elizabeth Hall, P. Gregory Hare, Christian Thomas, Rolf Mathewes, Catherine La Farge, André L. Martel, Peter D. Heintzman, and Beth Shapiro Abstract: A partial skeleton of a bison was recovered during residential house construction in Whitehorse, Yukon, Canada. The specimen represents a young (estimated 6 year old) bison individual that died, was partially scavenged by carnivores, and subsequently buried by calcareous silt sediment in a pond or small lake during the middle Holocene, ϳ5400 years ago. Palaeoenvironmental data, including molluscs, pollen, vascular plant, and bryophyte macrofossils demonstrate that the small waterbody was surrounded by white spruce dominated boreal forest. Morphometric analysis of the skeleton reveals that its taxonomic affinity is ambiguous, likely owing to it representing an ontogenetically young individual, though it does share some cranial and horn core characteristics of named species such as Bison occidentalis or Bison priscus. Mitochondrial genomic data confirm that this bison belongs to Clade 2A (northern clade), which represents Pleistocene steppe bison (B. cf. priscus) in Beringia through the Holocene and is not represented in living bison species. These data further demonstrate that northern steppe bison population survived the late Pleistocene extinction event, persisted locally in southern Yukon into the Holocene, and are best characterized as a species with a high degree of morphological variability and ecological flexibility. Résumé : Le squelette partiel d’un bison a été récupéré durant la construction d’immeubles résidentiels a` Whitehorse (Yukon, Canada). Le spécimen représente un unique jeune bison (d’âge estimé a` 6 ans) qui est mort, a été partiellement dévoré par des carnivores pour ensuite être enseveli sous du silt calcareux dans un étang ou un petit lac durant l’Holocène moyen, il y a ϳ5400 ans. -

Quaternary Records of the Dire Wolf, Canis Dirus, in North and South America
Quaternary records of the dire wolf, Canis dirus, in North and South America ROBERT G. DUNDAS Dundas, R. G. 1999 (September): Quaternary records of the dire wolf, Canis dirus, in North and South Ameri- ca. Boreas, Vol. 28, pp. 375–385. Oslo. ISSN 0300-9483. The dire wolf was an important large, late Pleistocene predator in North and South America, well adapted to preying on megaherbivores. Geographically widespread, Canis dirus is reported from 136 localities in North America from Alberta, Canada, southward and from three localities in South America (Muaco, Venezuela; Ta- lara, Peru; and Tarija, Bolivia). The species lived in a variety of environments, from forested mountains to open grasslands and plains ranging in elevation from sea level to 2255 m (7400 feet). Canis dirus is assigned to the Rancholabrean land mammal age of North America and the Lujanian land mammal age of South Amer- ica and was among the many large carnivores and megaherbivores that became extinct in North and South America near the end of the Pleistocene Epoch. Robert G. Dundas, Department of Geology, California State University, Fresno, California 93740-8031, USA. E-mail: [email protected]; received 20th May 1998, accepted 23rd March 1999 Because of the large number of Canis dirus localities Rancho La Brea, comparing them with Canis lupus and and individuals recovered from the fossil record, the dire wolf specimens from other localities. Although dire wolf is the most commonly occurring large knowledge of the animal’s biology had greatly predator in the Pleistocene of North America. By increased by 1912, little was known about its strati- contrast, the species is rare in South America. -

Frequency of Pathology in a Large Natural Sample from Natural Trap
Reumatismo, 2003; 55(1):58-65 RUBRICA DALLA RICERCA ALLA PRATICA Frequency of pathology in a large natural sample from Natural Trap Cave with special remarks on erosive disease in the Pleistocene La patologia osteoarticolare, con particolare riguardo a quella di tipo erosivo, nel Pleistocene: studio di un campione di reperti paleopatologici provenienti dalla Natural Trap Cave (Wyoming, USA) B.M. Rothschild1, L.D. Martin2 1The Arthritis Center of Northeast Ohio, University of Kansas Museum of Natural History, Carnegie Museum of Natural History, Northeastern Ohio Universities College of Medicine and University of Akron; 2Museum of Natural History and Department of Systematics and Ecology, University of Kansas, Lawrence, Kansas 66045 RIASSUNTO Nel presente studio vengono riportati i rilievi paleopatologici, con particolare riguardo alla presenza di artrite ero- siva, di osteoartrosi, di DISH , nonché ai segni di danno della dentizione, relativi ad un’ampia popolazione di mam- miferi, i cui resti (più di 30.000 ossa di 24 specie diverse) sono stati ritrovati nella Natural Trap Cave, Wyoming, USA. L’evidenza di artrite erosiva è confinata ai bovidi, Bison, Ovis e Bootherium, fatto osservabile anche in bisonti del tardo Pleistocene ritrovati nel Kansas (Twelve Mile Creek) e in un’altra località del Wisconsin, riferibile cronologi- camente al primo Olocene. Questi dati, ovvero la restrizione di tali segni di patologia articolare ad un singolo gene- re animale (Bovidi) e ad un determinato periodo storico, rende plausibile l’ipotesi che un agente patogeno, identifi- cabile col Mycobacterium tubercolosis, possa essere stato implicato nella genesi dell’artrite erosiva. Osteoartrosi e DISH sono risultate poco rappresentate nella popolazione animale della Natural Trap Cave, anche se il genere Bi- son ha dimostrato una discreta prevalenza di segni di osteoartrosi. -

The Holocene
The Holocene http://hol.sagepub.com The Holocene history of bighorn sheep (Ovis canadensis) in eastern Washington state, northwestern USA R. Lee Lyman The Holocene 2009; 19; 143 DOI: 10.1177/0959683608098958 The online version of this article can be found at: http://hol.sagepub.com/cgi/content/abstract/19/1/143 Published by: http://www.sagepublications.com Additional services and information for The Holocene can be found at: Email Alerts: http://hol.sagepub.com/cgi/alerts Subscriptions: http://hol.sagepub.com/subscriptions Reprints: http://www.sagepub.com/journalsReprints.nav Permissions: http://www.sagepub.co.uk/journalsPermissions.nav Citations http://hol.sagepub.com/cgi/content/refs/19/1/143 Downloaded from http://hol.sagepub.com at University of Missouri-Columbia on January 13, 2009 The Holocene 19,1 (2009) pp. 143–150 The Holocene history of bighorn sheep (Ovis canadensis) in eastern Washington state, northwestern USA R. Lee Lyman* (Department of Anthropology, 107 Swallow Hall, University of Missouri-Columbia, Columbia MO 65211, USA) Received 9 May 2008; revised manuscript accepted 30 June 2008 Abstract: Historical data are incomplete regarding the presence/absence and distribution of bighorn sheep (Ovis canadensis) in eastern Washington State. Palaeozoological (archaeological and palaeontological) data indicate bighorn were present in many areas there during most of the last 10 000 years. Bighorn occupied the xeric shrub-steppe habitats of the Channeled Scablands, likely because the Scablands provided the steep escape terrain bighorn prefer. The relative abundance of bighorn is greatest during climatically dry intervals and low during a moist period. Bighorn remains tend to increase in relative abundance over the last 6000 years. -

SFU Thesis Template Files
Reconstructing Northern Fur Seal Population Diversity through Ancient and Modern DNA Data by Cara Leanne Halseth B.Sc., University of Northern British Columbia, 2011 Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of Master of Arts in the Department of Archaeology Faculty of Environment © Cara Leanne Halseth 2015 SIMON FRASER UNIVERSITY Summer 2015 Approval Name: Cara Leanne Halseth Degree: Master of Arts Title: Reconstructing Northern Fur Seal Population Diversity through Ancient and Modern DNA Data Examining Committee: Chair: Catherine D‟Andrea Professor Dongya Yang Senior Supervisor Professor Deborah C. Merrett Supervisor Adjunct Professor Iain McKechnie External Examiner SSHRC Postdoctoral Fellowship Dept of Anthropology University of Oregon Date Defended/Approved: July 2, 2015 ii Abstract Archaeological and historic evidence suggests that northern fur seal (Callorhinus ursinus) has undergone several population and distribution changes (including commercial sealing) potentially resulting in a loss of genetic diversity and population structure. This study analyzes 36 unpublished mtDNA sequences from archaeological sites 1900-150 BP along the Pacific Northwest Coast from Moss et al. (2006) as well as published data (primarily Pinsky et al. [2010]) to investigate this species‟ genetic diversity and population genetics in the past. The D-loop data shows high nucleotide and haplotype diversity, with continuity of two separate subdivisions (haplogroups) through time. Nucleotide mismatch analysis suggests population expansion in both ancient and modern data. AMOVA analysis (FST and ΦST) reveals some „structure‟ detectable between several archaeological sites. While the data reviewed here did not reveal dramatic patterning, the AMOVA analysis does identify several significant FST values, indicating some level of ancient population „structure‟, which deserves future study. -

Reunion and Symposium the Past, Present, and Future of Ecology at Uga
SCHEDULE AT A GLANCE Jan. 12-14, 2018 REUNION AND SYMPOSIUM THE PAST, PRESENT, AND FUTURE OF ECOLOGY AT UGA FRIDAY, JAN. 12, 2018 4:00 – 7:00 p.m. “First” Friday Welcoming Reception (Ecology Building/Ecology Tent) The reunion symposium officially begins with a welcome reception featuring food, beverages, music and the opportunity to catch up with friends and classmates. (OK, technically it’s second Friday.) Registration and packet pick-up will be available in the Ecology lobby. Odum School Art Exhibit (Ecology Seminar Room) 5:30 – 6:00 p.m. Ecotones “UGA's ecologically-minded co-ed a cappella group” (Ecology Auditorium) 6:00 p.m. Slideshow of alumni and current & past faculty (Ecology Auditorium) SATURDAY, JAN. 13, 2018 7:30 – 8:30 a.m. Breakfast & Registration (Forestry Auditorium) Coffee and pastries available 8:30 – 10:00 a.m. Session I: Opening Plenary Session (Forestry Auditorium) • Welcome from DEAN JOHN GITTLEMAN • PAMELA WHITTEN | Senior Vice President for Academic Affairs and Provost • PETER RAVEN | President Emeritus of the Missouri Botanical Garden and former Home Secretary of the National Academy of Sciences Ecosystems and the Ecology of Change Plenary Address by Monica Turner, PhD ‘85 MONICA TURNER, a member of the National Academy of Sciences since 2004 and Past President of the Ecological Society of America, is a landscape ecologist who studies the ecosystem effects of fire and other disturbances and the ecological effects of climate and land use change. 10:00 – 10:15 a.m. Coffee Break (Ecology Tent) 10:15 – 11:30 a.m. Session II: Foundational Research and Current Connections: Alumni Perspectives (Forestry) • CHRISTOPHER D’ELIA, PhD ‘74 • WEIXIN CHENG, PhD ‘89 • EVELYN GAISER, PhD ‘97 • CHRISTINA FAUST, B.S./M.S., ‘09 11:30 – 12:00 Ecology and the University of Georgia Jere Morehead, President, University of Georgia p.m. -
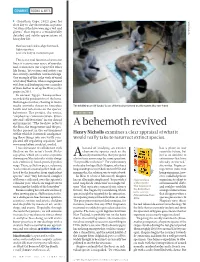
A Behemoth Revived
COMMENT BOOKS & ARTS (Jonathan Cape, 2012) goes for close day-to-day observation, in poems “written at the hive wearing a veil and gloves” that express a wonderfully detailed and subtle appreciation of RIA NOVOSTI/SPL honeybee life: How bees touch and re-align their touch. Light migration; noise of a body in continual repair This is one vital function of art in our lives: it restores our sense of wonder, and so increases our respect for other life forms. Yet writers and artists can also actively contribute new knowledge. One example of this is the work of visual artist Amy Shelton, whose engagement with bees and beekeeping over a number of years led her to set up the Honeyscribe project in 2011. In ancient Egypt, ‘honeyscribes’ recorded the productivity of the hives. Shelton goes further, charting in multi media artworks threats to honeybee The 40,000-year-old ‘Lyuba’ is one of the best-preserved woolly mammoths ever found. health and reflections on the species’ behaviour. Her project, she writes, DE-EXTINCTION “emphasizes communication, diver sity and collaboration” in our shared environment. “The beehive reflects the flora, the temperature and the pes A behemoth revived ticides present in the environment within which it is situated, amalgamat Henry Nicholls examines a clear appraisal of what it ing these things into one vastly com would really take to resurrect extinct species. plicated self-regulating organism” (see www.amyshelton.co.uk/art_works). I was fortunate to collaborate with hazard of studying an extinct has a place in our Shelton on the artist’s book Melis- charismatic species such as the scientific future, but sographia, which sets a series of poems woolly mammoth is that you spend not as an antidote to drawing on Maeterlinck’s study along Aa lot of time answering the same question: extinctions that have side embossed, hand-painted pollen “Is it possible to clone it?” For evolutionary already occurred,” maps. -

Small Mammal Faunal Stasis in Natural Trap Cave (Pleistocene– Holocene), Bighorn Mountains, Wyoming
SMALL MAMMAL FAUNAL STASIS IN NATURAL TRAP CAVE (PLEISTOCENE– HOLOCENE), BIGHORN MOUNTAINS, WYOMING BY C2009 Daniel R. Williams Submitted to the graduate degree program in Ecology and Evolutionary Biology and the Graduate Faculty of the University of Kansas in partial fulfillment of the requirements for the degree of Doctor of Philosophy. ______________________ Larry D. Martin/Chairperson Committee members* _____________________* Bruce S. Lieberman _____________________* Robert M. Timm _____________________* Bryan L. Foster _____________________* William C. Johnson Date defended: April 21, 2009 ii The Dissertation Committee for Daniel Williams certifies that this is the approved version of the following dissertation: SMALL MAMMAL FAUNAL STASIS IN NATURAL TRAP CAVE (PLEISTOCENE– HOLOCENE), BIGHORN MOUNTAINS, WYOMING Commmittee: ____________________________________ Larry D. Martin/Chairperson* ____________________________________ Bruce S. Lieberman ____________________________________ Robert M. Timm ____________________________________ Bryan L. Foster ____________________________________ William C. Johnson Date approved: April, 29, 2009 iii ABSTRACT Paleocommunity behavior through time is a topic of fierce debate in paleoecology, one with ramifications for the general study of macroevolution. The predominant viewpoint is that communities are ephemeral objects during the Quaternary that easily fall apart, but evidence exists that suggests geography and spatial scale plays a role. Natural Trap Cave is a prime testing ground for observing how paleocommunities react to large-scale climate change. Natural Trap Cave has a continuous faunal record (100 ka–recent) that spans the last glacial cycle, large portions of which are replicated in local rockshelters, which is used here to test for local causes of stasis. The Quaternary fauna of North America is relatively well sampled and dated, so the influence of spatial scale and biogeography on local community change can also be tested for. -

A Middle Holocene Steppe Bison and Paleoenvironments from the Versleuce Meadows, Whitehorse, Yukon, Canada Grant D
1138 ARTICLE A middle Holocene steppe bison and paleoenvironments from the Versleuce Meadows, Whitehorse, Yukon, Canada Grant D. Zazula, Elizabeth Hall, P. Gregory Hare, Christian Thomas, Rolf Mathewes, Catherine La Farge, André L. Martel, Peter D. Heintzman, and Beth Shapiro Abstract: A partial skeleton of a bison was recovered during residential house construction in Whitehorse, Yukon, Canada. The specimen represents a young (estimated 6 year old) bison individual that died, was partially scavenged by carnivores, and subsequently buried by calcareous silt sediment in a pond or small lake during the middle Holocene, ϳ5400 years ago. Palaeoenvironmental data, including molluscs, pollen, vascular plant, and bryophyte macrofossils demonstrate that the small waterbody was surrounded by white spruce dominated boreal forest. Morphometric analysis of the skeleton reveals that its taxonomic affinity is ambiguous, likely owing to it representing an ontogenetically young individual, though it does share some cranial and horn core characteristics of named species such as Bison occidentalis or Bison priscus. Mitochondrial genomic data confirm that this bison belongs to Clade 2A (northern clade), which represents Pleistocene steppe bison (B. cf. priscus) in Beringia through the Holocene and is not represented in living bison species. These data further demonstrate that northern steppe bison population survived the late Pleistocene extinction event, persisted locally in southern Yukon into the Holocene, and are best characterized as a species with a high degree of morphological variability and ecological flexibility. Résumé : Le squelette partiel d’un bison a été récupéré durant la construction d’immeubles résidentiels a` Whitehorse (Yukon, Canada). Le spécimen représente un unique jeune bison (d’âge estimé a` 6 ans) qui est mort, a été partiellement dévoré par des carnivores pour ensuite être enseveli sous du silt calcareux dans un étang ou un petit lac durant l’Holocène moyen, il y a ϳ5400 ans.