Perspectives in Comparative Genomics and Evolution Meeting Report
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Horseshoe-Based Bayesian Nonparametric Estimation of Effective Population Size Trajectories
Received: 13 August 2018 Accepted: 9 July 2019 DOI: 10.1111/biom.13276 BIOMETRIC METHODOLOGY Horseshoe-based Bayesian nonparametric estimation of effective population size trajectories James R. Faulkner1,2 Andrew F. Magee3 Beth Shapiro4,5 Vladimir N. Minin6 1Quantitative Ecology and Resource Abstract Management, University of Washington, Seattle, Washington Phylodynamics is an area of population genetics that uses genetic sequence data to 2Fish Ecology Division, Northwest Fisheries estimate past population dynamics. Modern state-of-the-art Bayesian nonparamet- Science Center, National Marine Fisheries ric methods for recovering population size trajectories of unknown form use either Service, NOAA, Seattle, Washington change-point models or Gaussian process priors. Change-point models suffer from 3Department of Biology, University of Washington, Seattle, Washington computational issues when the number of change-points is unknown and needs to 4Ecology and Evolutionary Biology be estimated. Gaussian process-based methods lack local adaptivity and cannot accu- Department and Genomics Institute, rately recover trajectories that exhibit features such as abrupt changes in trend or vary- University of California Santa Cruz, Santa Cruz, California ing levels of smoothness. We propose a novel, locally adaptive approach to Bayesian 5Howard Hughes Medical Institute, nonparametric phylodynamic inference that has the flexibility to accommodate a University of California Santa Cruz, large class of functional behaviors. Local adaptivity results from modeling the log- Santa Cruz, California transformed effective population size a priori as a horseshoe Markov random field, 6Department of Statistics, University of arecentlyproposedstatisticalmodelthatblendstogetherthebestpropertiesofthe California Irvine, Irvine, California change-point and Gaussian process modeling paradigms. We use simulated data to Correspondence assess model performance, and find that our proposed method results in reduced bias Vladimir N. -

Division Or Research Center Department Faculty Description
Division or Research Center Include in 2019 Department Faculty Description Sust. Research Reason for excluding (Y/N/M) Anderson's current research incorporates computer technologies to engage questions Y about land use and social interventions into the environment. His recent work, Silicon Monuments - in collaboration with the Silicon Valley Toxics Coalition - uses augmented reality software on hand-held devices to create a site-specific, multimedia documentary about toxic Superfund sites in Silicon Valley. Viewers can explore the sites and interact with the documentary, which reveals hidden environmental damage and its health and social costs. Website link: http://arts.ucsc.edu/faculty/eanderson/ Arts Art Elliott W. Anderson A. Laurie Palmer’s work is concerned with material explorations of matter’s active Y nature as it asserts itself on different scales and in different speeds, and with collaborating on strategic actions in the contexts of social and environmental justice. These two directions sometimes run parallel and sometimes converge, taking form as sculpture, installation, writing, and public projects. Collaboration, with other humans and with non-humans, is a central ethic in her practice. Website link: http: //alauriepalmer.net/ Arts Art Laurie Palmer Contemporary art and visual culture, investigating in particular the diverse ways that Y artists and activists have negotiated crises associated with globalization, including the emerging conjunction of post-9/11 political sovereignty and statelessness, the hauntings of the colonial past, and the growing biopolitical conflicts around ecology and climate change. Most recently Demos is the author of Decolonizing Nature: Contemporary Art and the Politics of Ecology (Sternberg Press, 2016), which investigates how concern for ecological crisis has entered the field of contemporary art and visual culture in recent years, and considers art and visual cultural practices globally. -

SFU Thesis Template Files
Reconstructing Northern Fur Seal Population Diversity through Ancient and Modern DNA Data by Cara Leanne Halseth B.Sc., University of Northern British Columbia, 2011 Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of Master of Arts in the Department of Archaeology Faculty of Environment © Cara Leanne Halseth 2015 SIMON FRASER UNIVERSITY Summer 2015 Approval Name: Cara Leanne Halseth Degree: Master of Arts Title: Reconstructing Northern Fur Seal Population Diversity through Ancient and Modern DNA Data Examining Committee: Chair: Catherine D‟Andrea Professor Dongya Yang Senior Supervisor Professor Deborah C. Merrett Supervisor Adjunct Professor Iain McKechnie External Examiner SSHRC Postdoctoral Fellowship Dept of Anthropology University of Oregon Date Defended/Approved: July 2, 2015 ii Abstract Archaeological and historic evidence suggests that northern fur seal (Callorhinus ursinus) has undergone several population and distribution changes (including commercial sealing) potentially resulting in a loss of genetic diversity and population structure. This study analyzes 36 unpublished mtDNA sequences from archaeological sites 1900-150 BP along the Pacific Northwest Coast from Moss et al. (2006) as well as published data (primarily Pinsky et al. [2010]) to investigate this species‟ genetic diversity and population genetics in the past. The D-loop data shows high nucleotide and haplotype diversity, with continuity of two separate subdivisions (haplogroups) through time. Nucleotide mismatch analysis suggests population expansion in both ancient and modern data. AMOVA analysis (FST and ΦST) reveals some „structure‟ detectable between several archaeological sites. While the data reviewed here did not reveal dramatic patterning, the AMOVA analysis does identify several significant FST values, indicating some level of ancient population „structure‟, which deserves future study. -

Model-Based Integration of Genomics and Metabolomics Reveals SNP Functionality in Mycobacterium Tuberculosis
Model-based integration of genomics and metabolomics reveals SNP functionality in Mycobacterium tuberculosis Ove Øyåsa,b,1, Sonia Borrellc,d,1, Andrej Traunerc,d,1, Michael Zimmermanne, Julia Feldmannc,d, Thomas Liphardta,b, Sebastien Gagneuxc,d, Jörg Stellinga,b, Uwe Sauere, and Mattia Zampierie,2 aDepartment of Biosystems Science and Engineering, ETH Zurich, 4058 Basel, Switzerland; bSIB Swiss Institute of Bioinformatics, 1015 Lausanne, Switzerland; cDepartment of Medical Parasitoloy and Infection Biology, Swiss Tropical and Public Health Institute, 4051 Basel, Switzerland; dUniversity of Basel, 4058 Basel, Switzerland; and eInstitute of Molecular Systems Biology, ETH Zurich, 8093 Zurich, Switzerland Edited by Ralph R. Isberg, Tufts University School of Medicine, Boston, MA, and approved March 2, 2020 (received for review September 12, 2019) Human tuberculosis is caused by members of the Mycobacterium infection of macrophages (29–32). Beyond analyses of individual tuberculosis complex (MTBC) that vary in virulence and transmis- laboratory strains, however, no systematic characterization and sibility. While genome-wide association studies have uncovered comparative analysis of intrinsic metabolic differences across several mutations conferring drug resistance, much less is known human-adapted MTBC clinical strains has been performed. about the factors underlying other bacterial phenotypes. Variation If the metabolic and other phenotypic diversity between in the outcome of tuberculosis infection and diseases has been MTBC strains contributes to and modulates pathogenicity, an attributed primarily to patient and environmental factors, but obvious question is: Which elements of the limited genetic di- recent evidence indicates an additional role for the genetic diver- versity in the MTBC are responsible for phenotypic strain di- sity among MTBC clinical strains. -

Reunion and Symposium the Past, Present, and Future of Ecology at Uga
SCHEDULE AT A GLANCE Jan. 12-14, 2018 REUNION AND SYMPOSIUM THE PAST, PRESENT, AND FUTURE OF ECOLOGY AT UGA FRIDAY, JAN. 12, 2018 4:00 – 7:00 p.m. “First” Friday Welcoming Reception (Ecology Building/Ecology Tent) The reunion symposium officially begins with a welcome reception featuring food, beverages, music and the opportunity to catch up with friends and classmates. (OK, technically it’s second Friday.) Registration and packet pick-up will be available in the Ecology lobby. Odum School Art Exhibit (Ecology Seminar Room) 5:30 – 6:00 p.m. Ecotones “UGA's ecologically-minded co-ed a cappella group” (Ecology Auditorium) 6:00 p.m. Slideshow of alumni and current & past faculty (Ecology Auditorium) SATURDAY, JAN. 13, 2018 7:30 – 8:30 a.m. Breakfast & Registration (Forestry Auditorium) Coffee and pastries available 8:30 – 10:00 a.m. Session I: Opening Plenary Session (Forestry Auditorium) • Welcome from DEAN JOHN GITTLEMAN • PAMELA WHITTEN | Senior Vice President for Academic Affairs and Provost • PETER RAVEN | President Emeritus of the Missouri Botanical Garden and former Home Secretary of the National Academy of Sciences Ecosystems and the Ecology of Change Plenary Address by Monica Turner, PhD ‘85 MONICA TURNER, a member of the National Academy of Sciences since 2004 and Past President of the Ecological Society of America, is a landscape ecologist who studies the ecosystem effects of fire and other disturbances and the ecological effects of climate and land use change. 10:00 – 10:15 a.m. Coffee Break (Ecology Tent) 10:15 – 11:30 a.m. Session II: Foundational Research and Current Connections: Alumni Perspectives (Forestry) • CHRISTOPHER D’ELIA, PhD ‘74 • WEIXIN CHENG, PhD ‘89 • EVELYN GAISER, PhD ‘97 • CHRISTINA FAUST, B.S./M.S., ‘09 11:30 – 12:00 Ecology and the University of Georgia Jere Morehead, President, University of Georgia p.m. -
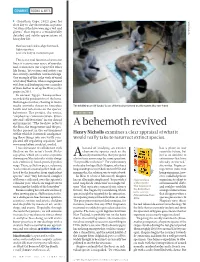
A Behemoth Revived
COMMENT BOOKS & ARTS (Jonathan Cape, 2012) goes for close day-to-day observation, in poems “written at the hive wearing a veil and gloves” that express a wonderfully detailed and subtle appreciation of RIA NOVOSTI/SPL honeybee life: How bees touch and re-align their touch. Light migration; noise of a body in continual repair This is one vital function of art in our lives: it restores our sense of wonder, and so increases our respect for other life forms. Yet writers and artists can also actively contribute new knowledge. One example of this is the work of visual artist Amy Shelton, whose engagement with bees and beekeeping over a number of years led her to set up the Honeyscribe project in 2011. In ancient Egypt, ‘honeyscribes’ recorded the productivity of the hives. Shelton goes further, charting in multi media artworks threats to honeybee The 40,000-year-old ‘Lyuba’ is one of the best-preserved woolly mammoths ever found. health and reflections on the species’ behaviour. Her project, she writes, DE-EXTINCTION “emphasizes communication, diver sity and collaboration” in our shared environment. “The beehive reflects the flora, the temperature and the pes A behemoth revived ticides present in the environment within which it is situated, amalgamat Henry Nicholls examines a clear appraisal of what it ing these things into one vastly com would really take to resurrect extinct species. plicated self-regulating organism” (see www.amyshelton.co.uk/art_works). I was fortunate to collaborate with hazard of studying an extinct has a place in our Shelton on the artist’s book Melis- charismatic species such as the scientific future, but sographia, which sets a series of poems woolly mammoth is that you spend not as an antidote to drawing on Maeterlinck’s study along Aa lot of time answering the same question: extinctions that have side embossed, hand-painted pollen “Is it possible to clone it?” For evolutionary already occurred,” maps. -

6.047/6.878 Lecture 4: Comparative Genomics I: Genome Annotation Using Evolutionary Signatures
6.047/6.878 Lecture 4: Comparative Genomics I: Genome Annotation Using Evolutionary Signatures Mark Smith (Partially adapted from notes by: Angela Yen, Christopher Robde, Timo Somervuo and Saba Gul) 9/18/12 1 Contents 1 Introduction 4 1.1 Motivation and Challenge......................................4 1.2 Importance of many closely{related genomes...........................4 2 Conservation of genomic sequences5 2.1 Functional elements in Drosophila .................................5 2.2 Rates and patterns of selection...................................5 3 Excess Constraint 6 3.1 Causes of Excess Constraint.....................................7 3.2 Modeling Excess Constraint.....................................8 3.3 Excess Constraint in the Human Genome.............................9 3.4 Examples of Excess Constraint................................... 10 4 Diversity of evolutionary signatures: An Overview of Selection Patterns 10 4.1 Selective Pressures On Different Functional Elements....................... 11 5 Protein{Coding Signatures 12 5.1 Reading{Frame Conservation (RFC)................................ 13 5.2 Codon{Substitution Frequencies (CSFs).............................. 14 5.3 Classification of Drosophila Genome Sequences.......................... 16 5.4 Leaky Stop Codons.......................................... 17 6 microRNA (miRNA) genes 19 6.1 Computational Challenge...................................... 20 6.2 Unusual miRNA Genes........................................ 21 7 Regulatory Motifs 22 7.1 Computationally Detecting -

Career Pathway Tracker 35 Years of Supporting Early Career Research Fellows Contents
Career pathway tracker 35 years of supporting early career research fellows Contents President’s foreword 4 Introduction 6 Scientific achievements 8 Career achievements 14 Leadership 20 Commercialisation 24 Public engagement 28 Policy contribution 32 How have the fellowships supported our alumni? 36 Who have we supported? 40 Where are they now? 44 Research Fellowship to Fellow 48 Cover image: Graphene © Vertigo3d CAREER PATHWAY TRACKER 3 President’s foreword The Royal Society exists to encourage the development and use of Very strong themes emerge from the survey About this report science for the benefit of humanity. One of the main ways we do that about why alumni felt they benefited. The freedom they had to pursue the research they This report is based on the first is by investing in outstanding scientists, people who are pushing the wanted to do because of the independence Career Pathway Tracker of the alumni of University Research Fellowships boundaries of our understanding of ourselves and the world around the schemes afford is foremost in the minds of respondents. The stability of funding and and Dorothy Hodgkin Fellowships. This us and applying that understanding to improve lives. flexibility are also highly valued. study was commissioned by the Royal Society in 2017 and delivered by the Above Thirty-five years ago, the Royal Society The vast majority of alumni who responded The Royal Society has long believed in the Careers Research & Advisory Centre Venki Ramakrishnan, (CRAC), supported by the Institute for President of the introduced our University Research Fellowships to the survey – 95% of University Research importance of identifying and nurturing the Royal Society. -

The Distribution and Evolution of Arabidopsis Thaliana Cis Natural Antisense Transcripts Johnathan Bouchard, Carlos Oliver and Paul M Harrison*
Bouchard et al. BMC Genomics (2015)16:444 DOI 10.1186/s12864-015-1587-0 RESEARCH ARTICLE Open Access The distribution and evolution of Arabidopsis thaliana cis natural antisense transcripts Johnathan Bouchard, Carlos Oliver and Paul M Harrison* Abstract Background: Natural antisense transcripts (NATs) are regulatory RNAs that contain sequence complementary to other RNAs, these other RNAs usually being messenger RNAs. In eukaryotic genomes, cis-NATs overlap the gene they complement. Results: Here, our goal is to analyze the distribution and evolutionary conservation of cis-NATs for a variety of available data sets for Arabidopsis thaliana, to gain insights into cis-NAT functional mechanisms and their significance. Cis-NATs derived from traditional sequencing are largely validated by other data sets, although different cis-NAT data sets have different prevalent cis-NAT topologies with respect to overlapping protein-coding genes. A. thaliana cis-NATs have substantial conservation (28-35% in the three substantive data sets analyzed) of expression in A. lyrata. We examined evolutionary sequence conservation at cis-NAT loci in Arabidopsis thaliana across nine sequenced Brassicaceae species (picked for optimal discernment of purifying selection), focussing on the parts of their sequences not overlapping protein- coding transcripts (dubbed ‘NOLPs’). We found significant NOLP sequence conservation for 28-34% NATs across different cis-NAT sets. This NAT NOLP sequence conservation versus A. lyrata is generally significantly correlated with conservation of expression. We discover a significant enrichment of transcription factor binding sites (as evidenced by CHIP-seq data) in NOLPs compared to randomly sampled near-gene NOLP-like DNA , that is linked to significant sequence conservation. -

Bayesian Phylogenetic and Phylodynamic Data Integration Using BEAST 1.10 Marc A
Virus Evolution, 2018, 4(1): vey016 doi: 10.1093/ve/vey016 Resources Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10 Marc A. Suchard,1,2,3,*,† Philippe Lemey,4,‡ Guy Baele,4,§ Daniel L. Ayres,5 Alexei J. Drummond,6,7,* and Andrew Rambaut8,*,** 1Department of Biomathematics, David Geffen School of Medicine, University of California, Los Angeles, 621 Charles E. Young Dr., South, Los Angeles, CA, 90095 USA, 2Department of Biostatistics, Fielding School of Public Health, University of California, Los Angeles, 650 Charles E, Young Dr., South, Los Angeles, CA, 90095 USA, 3Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, 695 Charles E. Young Dr., South, Los Angeles, CA, 90095 USA, 4Department of Microbiology and Immunology, Rega Institute, KU Leuven, Herestraat 49, 3000 Leuven, Belgium, 5Center for Bioinformatics and Computational Biology, University of Maryland, College Park, 125 Biomolecular Science Bldg #296, College Park, MD 20742 USA, , 6Department of Computer Science, University of Auckland, 303/38 Princes St., Auckland, 1010 NZ, 7Centre for Computational Evolution, University of Auckland, 303/38 Princes St., Auckland, 1010 NZ and 8Institute of Evolutionary Biology, University of Edinburgh, Ashworth Laboratories, Edinburgh, EH9 3FL UK *Corresponding author: E-mail: [email protected] (M.A.S.); [email protected] (A.J.D.); [email protected] (A.R.) †http://orcid.org/0000-0001-9818-479X ‡http://orcid.org/0000-0003-2826-5353 §http://orcid.org/0000-0002-1915-7732 **http://orcid.org/0000-0003-4337-3707 Abstract The Bayesian Evolutionary Analysis by Sampling Trees (BEAST) software package has become a primary tool for Bayesian phylogenetic and phylodynamic inference from genetic sequence data. -

A New Genus of Horse from Pleistocene North America
RESEARCH ARTICLE A new genus of horse from Pleistocene North America Peter D Heintzman1,2*, Grant D Zazula3, Ross DE MacPhee4, Eric Scott5,6, James A Cahill1, Brianna K McHorse7, Joshua D Kapp1, Mathias Stiller1,8, Matthew J Wooller9,10, Ludovic Orlando11,12, John Southon13, Duane G Froese14, Beth Shapiro1,15* 1Department of Ecology and Evolutionary Biology, University of California, Santa Cruz, Santa Cruz, United States; 2Tromsø University Museum, UiT - The Arctic University of Norway, Tromsø, Norway; 3Yukon Palaeontology Program, Government of Yukon, Whitehorse, Canada; 4Department of Mammalogy, Division of Vertebrate Zoology, American Museum of Natural History, New York, United States; 5Cogstone Resource Management, Incorporated, Riverside, United States; 6California State University San Bernardino, San Bernardino, United States; 7Department of Organismal and Evolutionary Biology, Harvard University, Cambridge, United States; 8Department of Translational Skin Cancer Research, German Consortium for Translational Cancer Research, Essen, Germany; 9College of Fisheries and Ocean Sciences, University of Alaska Fairbanks, Fairbanks, United States; 10Alaska Stable Isotope Facility, Water and Environmental Research Center, University of Alaska Fairbanks, Fairbanks, United States; 11Centre for GeoGenetics, Natural History Museum of Denmark, København K, Denmark; 12Universite´ Paul Sabatier, Universite´ de Toulouse, Toulouse, France; 13Keck-CCAMS Group, Earth System Science Department, University of California, Irvine, Irvine, United States; 14Department of Earth and Atmospheric Sciences, University of Alberta, Edmonton, Canada; 15UCSC Genomics Institute, University of California, Santa Cruz, Santa Cruz, United States *For correspondence: [email protected] (PDH); [email protected] (BS) Competing interests: The Abstract The extinct ‘New World stilt-legged’, or NWSL, equids constitute a perplexing group authors declare that no of Pleistocene horses endemic to North America. -

Transcriptional Interferences in Cis Natural Antisense Transcripts of Humans and Mice
Copyright Ó 2007 by the Genetics Society of America DOI: 10.1534/genetics.106.069484 Transcriptional Interferences in cis Natural Antisense Transcripts of Humans and Mice Naoki Osato, Yoshiyuki Suzuki, Kazuho Ikeo and Takashi Gojobori1 Center for Information Biology and DNA Data Bank of Japan, National Institute of Genetics, Research Organization of Information and Systems, Mishima 411-8540, Japan Manuscript received December 9, 2006 Accepted for publication March 21, 2007 ABSTRACT For a significant fraction of mRNAs, their expression is regulated by other RNAs, including cis natural antisense transcripts (cis -NATs) that are complementary mRNAs transcribed from opposite strands of DNA at the same genomic locus. The regulatory mechanism of mRNA expression by cis -NATs is unknown, although a few possible explanations have been proposed. To understand this regulatory mechanism, we conducted a large-scale analysis of the currently available data and examined how the overlapping arrange- ments of cis -NATs affect their expression level. Here, we show that for both human and mouse the expression level of cis -NATs decreases as the length of the overlapping region increases. In particular, the proportions of the highly expressed cis -NATs in all cis -NATs examined were 36 and 47% for human and mouse, respectively, when the overlapping region was ,200 bp. However, both proportions decreased to virtually zero when the overlapping regions were .2000 bp in length. Moreover, the distribution of the expression level of cis -NATs changes according to different types of the overlapping pattern of cis -NATs in the genome. These results are consistent with the transcriptional collision model for the regulatory mechanism of gene expression by cis -NATs.