Contrasted Patterns in Mating-Type Chromosomes in Fungi: Hotspots
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evolution of Genetic Systems in Filamentous Ascomycetes
Evolution of Genetic Systems in Filamentous Ascomycetes Evolutie van genetische systemen in hyphenvormende zakjeszwammen 0000 0513 3836 Promotor: dr. R.F. Hoekstra hoogleraar in de populatie- en kwantitatieve genetica fjtfoiißi f ßin Maarten J. Nauta Evolution of Genetic Systems in Filamentous Ascomycetes Proefschrift ter verkrijging van de graad van doctor in de landbouw- en milieuwetenschappen op gezag van de rector magnificus, dr. C.M. Karssen, in het openbaar te verdedigen op woensdag 12januar i 1994 des namiddags te vier uur in de Aula van de Landbouwuniversiteit te Wageningen. 15 0 S(p^ZJ> These investigations were supported by the Netherlands Organization for Scientific Research (N.W.O.). BibUt/FHEEK LAMDbOirWUNIVERSITEJi. WAGE NINGE N CIP-GEGEVENS KONINKLIJKE BIBLIOTHEEK, DEN HAAG Nauta, Maarten J. Evolution of genetic systems in filamentous ascomycetes / Maarten J. Nauta. - [ S.l. : s.n.]. -111 . Thesis Wageningen. - With ref. - With summary in Dutch. ISBN 90-5485-199-6 Subject headings: population genetics / ascomycetes. omslagontwerp: Ernst van Cleef foto omslag: Barrages tussen verschillende stammen van Podospora anserina als gevolg van vegetatieve incompatibiliteit. (met dank aan Inge Haspels) aan mijn ouders Voorwoord Dit proefschrift is het resultaat van vier jaar onderzoek, verricht bij de vakgroep Erfelijkheidsleer van de Landbouwuniversiteit in Wageningen. In zekere zin valt zo'n proefschrift te vergelijken met een levend wezen. Uit de genetica is bekend dat de verschijningsvorm van elk levend wezen tot stand komt door een combinatie van erfelijke aanleg en invloeden uit de omgeving. Voor een proefschrift geldt eigenlijk hetzelfde: Zowel het werk van de auteur, als de bijdragen van zijn omgeving zijn onontbeerlijk om tot een verschijningsvorm te komen. -

Mechanisms Ofcarcinogenesis
Proc. Natl. Acad. Sci. USA Vol. 75, No. 12, pp. 6149-6153, December 1978 Genetics Tumor promoter induces sister chromatid exchanges: Relevance to mechanisms of carcinogenesis (12-0-tetradecanoylphorbol 13-acetate/bromodeoxyuridine labeling/recombination/mitotic segregation/recessive mutations) ANNE R. KINSELLA AND MIROSLAV RADMAN D6partement de Biologie Moleculaire, Universite Libre de Bruxelles, B 1640 Rhode-St-Genese, Belgium Communicated by J. D. Watson, August 31, 1978 ABSTRACT 12-0-Tetradecanoylphorbol 13-acetate (TPA), Six possible mechanisms for the segregation of a recessive a powerful tumor promoter, is shown to induce sister chromatid mutation from a heterozygous cell are presented in 1. exchanges (SCEs), whereas the nonpromoting derivative 4-0- Fig. (i) methyl-TPA does not. Inhibitors of tumor promotion-antipain, Chromosomal rearrangement or deletion and (ii) one-step leupeptin, and fluocinolone acetonide-inhibit formation of nondisjunction both lead to hemizygosity, while (iii) two-step such TPA-induced SCEs. TPA is a unique agent in its induction nondisjunction, (iv) aberrant mitotic segregation (in the absence of SCEs in the absence of DNA damage, chromosome aberra- of nondisjunction or mitotic recombination), (v) increase in tions, mutagenesis, or significant toxicity. Because TPA is ploidy plus chromosome loss, and (vi) mitotic recombination known to induce several gene functions, we speculate that it all lead to might also induce enzymes involved in genetic recombination. homozygosity. Only the events leading to homozy- Thus, the irreversible step in tumor promotion might be the re- gosity (iii-vi) are consistent with the observation that initiation sult of an aberrant mitotic segregation event leading to the ex- must precede promotion in order to produce an enhanced pression of carcinogen/mutagen-induced recessive genetic or carcinogenic effect. -

Algal Sex Determination and the Evolution of Anisogamy James Umen, Susana Coelho
Algal Sex Determination and the Evolution of Anisogamy James Umen, Susana Coelho To cite this version: James Umen, Susana Coelho. Algal Sex Determination and the Evolution of Anisogamy. Annual Review of Microbiology, Annual Reviews, 2019, 73 (1), 10.1146/annurev-micro-020518-120011. hal- 02187088 HAL Id: hal-02187088 https://hal.sorbonne-universite.fr/hal-02187088 Submitted on 17 Jul 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Annu. Rev. Microbiol. 2019. 73:X–X https://doi.org/10.1146/annurev-micro-020518-120011 Copyright © 2019 by Annual Reviews. All rights reserved Umen • Coelho www.annualreviews.org • Algal Sexes and Mating Systems Algal Sex Determination and the Evolution of Anisogamy James Umen1 and Susana Coelho2 1Donald Danforth Plant Science Center, St. Louis, Missouri 63132, USA; email: [email protected] 2Sorbonne Université, UPMC Université Paris 06, CNRS, Algal Genetics Group, UMR 8227, Integrative Biology of Marine Models, Station Biologique de Roscoff, CS 90074, F-29688, Roscoff, France [**AU: Please write the entire affiliation in French or write it all in English, rather than a combination of English and French**] ; email: [email protected] Abstract Algae are photosynthetic eukaryotes whose taxonomic breadth covers a range of life histories, degrees of cellular and developmental complexity, and diverse patterns of sexual reproduction. -

Prospects & Overviews Meiotic Versus Mitotic Recombination: Two Different
Prospects & Overviews Meiotic versus mitotic recombination: Two different routes for double-strand Review essays break repair The different functions of meiotic versus mitotic DSB repair are reflected in different pathway usage and different outcomes Sabrina L. Andersen1) and Jeff Sekelsky1)2)Ã Studies in the yeast Saccharomyces cerevisiae have vali- Introduction dated the major features of the double-strand break repair (DSBR) model as an accurate representation of The existence of DNA recombination was revealed by the behavior of segregating traits long before DNA was identified the pathway through which meiotic crossovers (COs) are as the bearer of genetic information. At the start of the 20th produced. This success has led to this model being century, pioneering Drosophila geneticists studied the behav- invoked to explain double-strand break (DSB) repair in ior of chromosomal ‘‘factors’’ that determined traits such as other contexts. However, most non-crossover (NCO) eye color, wing shape, and bristle length. In 1910 Thomas Hunt recombinants generated during S. cerevisiae meiosis do Morgan published the observation that the linkage relation- not arise via a DSBR pathway. Furthermore, it is becom- ships of these factors were shuffled during meiosis [1]. Building on this discovery, in 1913 A. H. Sturtevant used ing increasingly clear that DSBR is a minor pathway for linkage analysis to determine the order of factors (genes) recombinational repair of DSBs that occur in mitotically- on a chromosome, thus simultaneously establishing that proliferating cells and that the synthesis-dependent genes are located at discrete physical locations along chromo- strand annealing (SDSA) model appears to describe somes as well as originating the classic tool of genetic map- mitotic DSB repair more accurately. -

Increase in Mitotic Recombination in Diploid Cells of Aspergillus Nidulans in Response to Ethidium Bromide
Genetics and Molecular Biology, 26, 3, 381-385 (2003) Copyright by the Brazilian Society of Genetics. Printed in Brazil www.sbg.org.br Research Article Increase in mitotic recombination in diploid cells of Aspergillus nidulans in response to ethidium bromide Tânia C.A. Becker, Simone J.R. Chiuchetta, Francielle Baptista and Marialba A.A. de Castro-Prado Universidade Estadual de Maringá, Departamento de Genética e Biologia Celular Maringá, PR, Brazil. Abstract Ethidium bromide (EB) is an intercalating inhibitor of topoisomerase II and its activities are related to chemotherapeutic drugs used in anti-cancer treatments. EB promotes several genotoxic effects in exposed cells by stabilising the DNA-enzyme complex. The recombinagenic potential of EB was evaluated in our in vivo study by the loss of heterozygosity of nutritional markers in diploid Aspergillus nidulans cells through Homozygotization Index (HI). A DNA repair mutation, uvsZ and a chromosome duplication DP (II-I) were introduced in the genome of tested cells to obtain a sensitive system for the recombinagenesis detection. EB-treated diploid cells had HI values significantly greater than the control at both concentrations (4.0 x 10-3 and 5.0 x 10-3 µM). Results indicate that the intercalating agent is potentially capable of inducing mitotic crossing-over in diploid A. nidulans cells. Key words: antineoplastic drug, Aspergillus nidulans, ethidium bromide, intercalating agent, mitotic crossing over. Received: February 24, 2003; Accepted: May 29, 2003. Introduction amplification and transpositions, but also causes neoplasies DNA topoisomerase II (topo II) is a ubiquitous en- progression through loss of heterozygosity at tumor- zyme that regulates DNA topologic interconversion during suppressor loci. -

Classifications of Fungi
Chapter 24 | Fungi 675 Sexual Reproduction Sexual reproduction introduces genetic variation into a population of fungi. In fungi, sexual reproduction often occurs in response to adverse environmental conditions. During sexual reproduction, two mating types are produced. When both mating types are present in the same mycelium, it is called homothallic, or self-fertile. Heterothallic mycelia require two different, but compatible, mycelia to reproduce sexually. Although there are many variations in fungal sexual reproduction, all include the following three stages (Figure 24.8). First, during plasmogamy (literally, “marriage or union of cytoplasm”), two haploid cells fuse, leading to a dikaryotic stage where two haploid nuclei coexist in a single cell. During karyogamy (“nuclear marriage”), the haploid nuclei fuse to form a diploid zygote nucleus. Finally, meiosis takes place in the gametangia (singular, gametangium) organs, in which gametes of different mating types are generated. At this stage, spores are disseminated into the environment. Review the characteristics of fungi by visiting this interactive site (http://openstaxcollege.org/l/ fungi_kingdom) from Wisconsin-online. 24.2 | Classifications of Fungi By the end of this section, you will be able to do the following: • Identify fungi and place them into the five major phyla according to current classification • Describe each phylum in terms of major representative species and patterns of reproduction The kingdom Fungi contains five major phyla that were established according to their mode of sexual reproduction or using molecular data. Polyphyletic, unrelated fungi that reproduce without a sexual cycle, were once placed for convenience in a sixth group, the Deuteromycota, called a “form phylum,” because superficially they appeared to be similar. -

Self-Fertility and Uni-Directional Mating-Type Switching in Ceratocystis Coerulescens, a Filamentous Ascomycete
Curr Genet (1997) 32: 52–59 © Springer-Verlag 1997 ORIGINAL PAPER T. C. Harrington · D. L. McNew Self-fertility and uni-directional mating-type switching in Ceratocystis coerulescens, a filamentous ascomycete Received: 6 July 1996 / 25 March 1997 Abstract Individual perithecia from selfings of most some filamentous ascomycetes. Although a switch in the Ceratocystis species produce both self-fertile and self- expression of mating-type is seen in these fungi, it is not sterile progeny, apparently due to uni-directional mating- clear if a physical movement of mating-type genes is in- type switching. In C. coerulescens, male-only mutants of volved. It is also not clear if the expressed mating-types otherwise hermaphroditic and self-fertile strains were self- of the respective self-fertile and self-sterile progeny are sterile and were used in crossings to demonstrate that this homologs of the mating-type genes in other strictly heter- species has two mating-types. Only MAT-2 strains are othallic species of ascomycetes. capable of selfing, and half of the progeny from a MAT-2 Sclerotinia trifoliorum and Chromocrea spinulosa show selfing are MAT-1. Male-only, MAT-2 mutants are self- a 1:1 segregation of self-fertile and self-sterile progeny in sterile and cross only with MAT-1 strains. Similarly, self- perithecia from selfings or crosses (Mathieson 1952; Uhm fertile strains generally cross with only MAT-1 strains. and Fujii 1983a, b). In tetrad analyses of selfings or crosses, MAT-1 strains only cross with MAT-2 strains and never self. half of the ascospores in an ascus are large and give rise to It is hypothesized that the switch in mating-type during self-fertile colonies, and the other ascospores are small and selfing is associated with a deletion of the MAT-2 gene. -

Sex in the Extremes: Lichen-Forming Fungi
Mycologist, Volume 19, Part 2 May 2005. ©Cambridge University Press Printed in the United Kingdom. DOI: 10.1017/S0269915XO5002016 Sex in the extremes: lichen-forming fungi FABIAN A. SEYMOUR, PETER D. CRITTENDEN & PAUL S. DYER* School of Biology, University of Nottingham, University Park, Nottingham, NG7 2RD, UK. Tel. +44 (0) 115 9513203, Fax +44 (0) 115 9513251 E-mail: [email protected]; [email protected] *Corresponding Author Lichens are characteristically found in environments subject to extremes of temperature, desiccation and low nutrient status. Despite this sexual structures are often formed in abundance. The underlying mechanisms of sex in lichen-forming fungi are discussed, together with possible ecological reasons for the persistence of sexuality. Special features of lichen sex are highlighted including sex at the limits of life on earth in Antarctica, re-licheniza- tion following sex and dispersal, and the perennial nature of lichen fruiting bodies. Keywords: lichen, fungi, sex, breeding system, (98%) belonging to the Ascomycotina (Kirk et al., symbiosis, extreme environments, Antarctica 2001). They display a variety of morphologies, from flattened crust (crustose) or leafy (foliose) forms to Lichens - living together in a long-term relation- shrubby or pendulous fruticose types (Honegger, 2001) ship (Figs 3, 4, 7, 8). Lichens are seen as a textbook example of a successful Life in extreme environments mutualistic symbiosis. They consist of at least two A key characteristic of lichens is that they have a organisms: a fungus (the ‘mycobiont’), and an remarkable ability to tolerate extreme environmental intimately associated photosynthetic partner (the conditions and sustain growth despite frequent cycles ‘photobiont’). -

Break-Induced Replication Occurs by Conservative DNA Synthesis
Break-induced replication occurs by conservative DNA synthesis Roberto A. Donnianni and Lorraine S. Symington1 Department of Microbiology and Immunology, Columbia University Medical Center, New York, NY 10032 Edited* by Thomas D. Petes, Duke University Medical Center, Durham, NC, and approved June 21, 2013 (received for review May 23, 2013) Break-induced replication (BIR) refers to recombination-dependent The mechanisms that limit the extent of DNA synthesis during DNA synthesis initiated from one end of a DNA double-strand gene conversion repair, but promote extensive DNA synthesis in break and can extend for more than 100 kb. BIR initiates by Rad51- BIR are poorly understood. Previous studies have shown that catalyzed strand invasion, but the mechanism for DNA synthesis is BIR can involve multiple rounds of strand invasion and disso- not known. Here, we used BrdU incorporation to track DNA ciation, suggesting repair might occur by progressive extension of synthesis during BIR and found that the newly synthesized strands the invading 3′ end until the chromosome terminus is reached, segregate with the broken chromosome, indicative of a conserva- with lagging strand synthesis initiating on the nascent displaced tive mode of DNA synthesis. Furthermore, we show the frequency strand (Fig. 1) (10, 15, 16). Nonetheless, the requirement for the of BIR is reduced and product formation is progressively delayed replicative minichromosome maintenance helicase and lagging when the donor is placed at an increasing distance from the strand synthesis components to detect early BIR intermediates telomere, consistent with replication by a migrating D-loop from suggests BIR might involve assembly of a replication fork for the site of initiation to the telomere. -

Perithecial Ascomycetes from the 400 Million Year Old Rhynie Chert: an Example of Ancestral Polymorphism
Mycologia, 97(1), 2005, pp. 269±285. q 2005 by The Mycological Society of America, Lawrence, KS 66044-8897 Perithecial ascomycetes from the 400 million year old Rhynie chert: an example of ancestral polymorphism Editor's note: Unfortunately, the plates for this article published in the December 2004 issue of Mycologia 96(6):1403±1419 were misprinted. This contribution includes the description of a new genus and a new species. The name of a new taxon of fossil plants must be accompanied by an illustration or ®gure showing the essential characters (ICBN, Art. 38.1). This requirement was not met in the previous printing, and as a result we are publishing the entire paper again to correct the error. We apologize to the authors. T.N. Taylor1 terpreted as the anamorph of the fungus. Conidioge- Department of Ecology and Evolutionary Biology, and nesis is thallic, basipetal and probably of the holoar- Natural History Museum and Biodiversity Research thric-type; arthrospores are cube-shaped. Some peri- Center, University of Kansas, Lawrence, Kansas thecia contain mycoparasites in the form of hyphae 66045 and thick-walled spores of various sizes. The structure H. Hass and morphology of the fossil fungus is compared H. Kerp with modern ascomycetes that produce perithecial as- Forschungsstelle fuÈr PalaÈobotanik, Westfalische cocarps, and characters that de®ne the fungus are Wilhelms-UniversitaÈt MuÈnster, Germany considered in the context of ascomycete phylogeny. M. Krings Key words: anamorph, arthrospores, ascomycete, Bayerische Staatssammlung fuÈr PalaÈontologie und ascospores, conidia, fossil fungi, Lower Devonian, my- Geologie, Richard-Wagner-Straûe 10, 80333 MuÈnchen, coparasite, perithecium, Rhynie chert, teleomorph Germany R.T. -
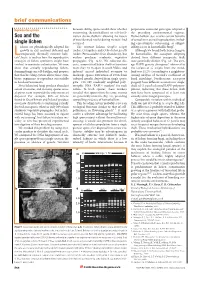
Brief Comms Layout 6/4Mx
brief communications Reproductive systems between sibling spores would show whether perpetuates successful genotypes adapted to outcrossing (heterothallism) or self-fertil- the prevailing environmental regimes. Sex and the ization (homothallism: allowing the fusion Homothallism also retains certain benefits of two identical nuclei during meiosis) had of sexual over asexual reproduction, includ- single lichen occurred. ing opportunistic outcrossing, as obligate ichens are physiologically adapted for The crustose lichens Graphis scripta selfing is rare in homothallic fungi8. growth in dry, nutrient-deficient and (order: Ostropales) and Ochrolechia parella Although we found both lichen fungi to Ltemporarily thermally extreme habi- (order: Pertusariales) fruit abundantly, but be homothallic, the ascospore offspring tats1, but it is unclear how the reproductive neither produces symbiotic vegetative derived from different conspecific thalli strategies of lichen symbionts might have propagules (Fig. 1a,b). We collected dis- were genetically distinct (Fig. 1d). The aver- evolved to maximize colonization. We now crete, symmetrical lichen thalli at locations age RAPD genetic divergence9 observed in show that sexually reproducing lichen- more than 10 m apart in south Wales, and five isolates of G. scripta from one wood- forming fungi can self-fertilize, and propose induced excised individual ascomata to land was 15.2% (according to a neighbour- that this breeding system allows these sym- discharge spores. Extraction of DNA from joining analysis of Jaccard’s coefficient of biotic organisms to reproduce successfully cultured mycelia derived from single spores band matching). Furthermore, ascospore in harsh environments. gave 218–263 randomly amplified poly- progeny from different ascomata on ‘single’ Most lichenized fungi produce abundant morphic DNA (RAPD) markers4 for each thalli of O. -

Mechanisms and Regulation of Mitotic Recombination in Saccharomyces Cerevisiae
YEASTBOOK GENOME ORGANIZATION AND INTEGRITY Mechanisms and Regulation of Mitotic Recombination in Saccharomyces cerevisiae Lorraine S. Symington,* Rodney Rothstein,† and Michael Lisby‡ *Department of Microbiology and Immunology, and yDepartment of Genetics and Development, Columbia University Medical Center, New York, New York 10032, and ‡Department of Biology, University of Copenhagen, DK-2200 Copenhagen, Denmark ABSTRACT Homology-dependent exchange of genetic information between DNA molecules has a profound impact on the maintenance of genome integrity by facilitating error-free DNA repair, replication, and chromosome segregation during cell division as well as programmed cell developmental events. This chapter will focus on homologous mitotic recombination in budding yeast Saccharomyces cerevisiae.However, there is an important link between mitotic and meiotic recombination (covered in the forthcoming chapter by Hunter et al. 2015) and many of the functions are evolutionarily conserved. Here we will discuss several models that have been proposed to explain the mechanism of mitotic recombination, the genes and proteins involved in various pathways, the genetic and physical assays used to discover and study these genes, and the roles of many of these proteins inside the cell. TABLE OF CONTENTS Abstract 795 I. Introduction 796 II. Mechanisms of Recombination 798 A. Models for DSB-initiated homologous recombination 798 DSB repair and synthesis-dependent strand annealing models 798 Break-induced replication 798 Single-strand annealing and microhomology-mediated end joining 799 B. Proteins involved in homologous recombination 800 DNA end resection 800 Homologous pairing and strand invasion 802 Rad51 mediators 803 Single-strand annealing 803 DNA translocases 804 DNA synthesis during HR 805 Resolution of recombination intermediates 805 III.