Molecular Identification of the Primary Root Fungal Endophytes of Dipodium Hamiltonianum (Orchidaceae)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ACT, Australian Capital Territory
Biodiversity Summary for NRM Regions Species List What is the summary for and where does it come from? This list has been produced by the Department of Sustainability, Environment, Water, Population and Communities (SEWPC) for the Natural Resource Management Spatial Information System. The list was produced using the AustralianAustralian Natural Natural Heritage Heritage Assessment Assessment Tool Tool (ANHAT), which analyses data from a range of plant and animal surveys and collections from across Australia to automatically generate a report for each NRM region. Data sources (Appendix 2) include national and state herbaria, museums, state governments, CSIRO, Birds Australia and a range of surveys conducted by or for DEWHA. For each family of plant and animal covered by ANHAT (Appendix 1), this document gives the number of species in the country and how many of them are found in the region. It also identifies species listed as Vulnerable, Critically Endangered, Endangered or Conservation Dependent under the EPBC Act. A biodiversity summary for this region is also available. For more information please see: www.environment.gov.au/heritage/anhat/index.html Limitations • ANHAT currently contains information on the distribution of over 30,000 Australian taxa. This includes all mammals, birds, reptiles, frogs and fish, 137 families of vascular plants (over 15,000 species) and a range of invertebrate groups. Groups notnot yet yet covered covered in inANHAT ANHAT are notnot included included in in the the list. list. • The data used come from authoritative sources, but they are not perfect. All species names have been confirmed as valid species names, but it is not possible to confirm all species locations. -

Australian Orchidaceae: Genera and Species (12/1/2004)
AUSTRALIAN ORCHID NAME INDEX (21/1/2008) by Mark A. Clements Centre for Plant Biodiversity Research/Australian National Herbarium GPO Box 1600 Canberra ACT 2601 Australia Corresponding author: [email protected] INTRODUCTION The Australian Orchid Name Index (AONI) provides the currently accepted scientific names, together with their synonyms, of all Australian orchids including those in external territories. The appropriate scientific name for each orchid taxon is based on data published in the scientific or historical literature, and/or from study of the relevant type specimens or illustrations and study of taxa as herbarium specimens, in the field or in the living state. Structure of the index: Genera and species are listed alphabetically. Accepted names for taxa are in bold, followed by the author(s), place and date of publication, details of the type(s), including where it is held and assessment of its status. The institution(s) where type specimen(s) are housed are recorded using the international codes for Herbaria (Appendix 1) as listed in Holmgren et al’s Index Herbariorum (1981) continuously updated, see [http://sciweb.nybg.org/science2/IndexHerbariorum.asp]. Citation of authors follows Brummit & Powell (1992) Authors of Plant Names; for book abbreviations, the standard is Taxonomic Literature, 2nd edn. (Stafleu & Cowan 1976-88; supplements, 1992-2000); and periodicals are abbreviated according to B-P- H/S (Bridson, 1992) [http://www.ipni.org/index.html]. Synonyms are provided with relevant information on place of publication and details of the type(s). They are indented and listed in chronological order under the accepted taxon name. Synonyms are also cross-referenced under genus. -

Fungal Endophytes in Australian Myco-Heterotrophic Orchids
Fungal Endophytes in Australian Myco-heterotrophic Orchids J.D.W. Dearnaley Centre for Systems Biology, The University of Southern Queensland, Toowoomba, Australia Summary The fungal endophytes of Australian myco-heterotrophic orchids are largely unknown. In this investigation we identified the fungal endophytes of three species of Australian myco-heterotrophic orchid: the terrestrial species Dipodium variegatum and Dipodium hamiltonianum and the vine-like Erythrorchis cassythoides. Similar to studies of myco-heterotrophic orchids in Nth America and Europe the fungal endophytes of Dipodium spp. are members of the Russulaceae. E. cassythoides contained both ectomycorrhizal and saprotrophic fungi suggesting that it can derive its carbon via fungi from both dead and living plant material. Introduction All orchids rely on fungi for seed germination and early growth. A number of orchid species are non-photosynthetic and are termed myco-heterotrophic because of their complete dependence on fungi throughout their life cycle. Molecular identification studies of fungal endophytes of myco-heterotrophic orchids have shown that these plants are colonized by higher basidiomycetes such as members of the Russulaceae and Thelophoraceae (Taylor and Bruns 1997, 1999, Girlanda et al. 2006) as well as members of the heterobasidomycete family Sebacinaceae (Selosse et al. 2002, Taylor et al. 2003). As these fungal groups contain ectomycorrhizal species it appears that myco-heterotrophic orchids are indirectly parasitic on the tree host partner of ectomycorrhizal partnerships. Evidence supporting this comes from McKendrick et al. (2000) who have shown that radiolabelled carbon can flow from tree to myco-hetero- trophic orchid via a fungal conduit, additionally, many myco-heterotrophic ©2006 by MEDIMOND S.r.l. -

WHAT DRIVES ORCHIDS.Pmd
J. Orchid Soc. India, 29: 23-30, 2015 ISSN 0971-5371 WHAT DRIVES ORCHIDS TOWARD MYCO-HETEROTROPHY? Julie Thakur, Akankankanksha Madan, Mayyyank D Dwivedi, and Prem L Uniyalalal Department of Botany, University of Delhi, Delhi-110 007, India Abstract Myco-heterotrophy describes the capability of orchids to obtain carbon from fungi. Depending upon varied durations of association with fungi, orchids have been categorized as those that require fungi during germination only as compared to those that are associated with fungi throughout the life span. It has always been the subject of interest to all the scientists to evaluate the mode of nutrition between this mutualistic association for which various modern approaches have been used. There are factors which led to evolution of autotrophy to initial or partial mycoheterotrophy and subsequently, to fully mycoheterotrophy. The present paper presents the overview of mycoheterotrophy, habitat, morphology, reproduction strategies, techniques, threats and conservation of mycoheterotrophy, and major opportunities for the future research. Introduction is divided into five sub-families: Apostasioideae, Vanilloideae, Cypripedioideae, Orchidoideae and MYCO-HETEROTROPHY IS a potential of a plant to Epidendroideae, of which Epidendroideae is the largest procure carbon from fungi. Leake (1994) coined the subfamily comprising of ca. 18,000 spp. and 650 term mycoheterotrophy (as myco-heterotrophy). To genera (Chase et al., 2015). As far as is known, all understand mycoheterotrophy, a basic understanding orchids depend on a myco-heterotrophic interaction about the mycorrhizal symbiosis is a pre-requisite. The with a symbiotic fungus for a part of their life cycle, term mycorrhiza is derived from the Greek words, especially for germination (Leake, 1994). -

Research Article Ultrastructural Studies and Molecular Characterization of Root-Associated Fungi of Crepidium Acuminatum (D
1 Research article Ultrastructural studies and molecular characterization of root-associated fungi of Crepidium acuminatum (D. Don) Szlach.: A threatened and medicinally important taxon JULIE THAKUR, MAYANK D. DWIVEDI# and PREM L. UNIYAL Department of Botany, University of Delhi, Delhi-110007 # For correspondence. E-mail: [email protected] Abstract Crepidium acuminatum (Orchidaceae) is a threatened medicinal orchid that grows under shady and moist forest floor where light remains for a very short period of time. Mycorrhizal association is known to be essential for seed germination and seedling establishment in majority of orchids. Identification of fungi that form mycorrhizae with orchids is of crucial importance for orchid conservation. We used both morphological as well as molecular approaches to study this plant-fungal interaction. Scanning electron microscopy showed that fungi grow and proliferate in the middle layers of cortex. Also, spiral root hairs were found along with roots hairs, which is an unusual observation. Spiral root hairs provide more surface area for fluid absorption and entrance of colonisers. Furthermore, total root genomic DNA was isolated and fungal ITS regions were PCR-amplified using specific primer combinations ITS1F/ITS4 and ITS1/ITS4tul. ITS sequences were obtained and analysed to know the closest sequence matches in the GenBank using BLASTn hosted by NLM-NCBI. Subject sequences were identified to be belonging to three main genera viz., Tulasnella, Aspergillus and Penicillium. Results indicate that mycorrhizal association is necessary for the growth and development of this plant. In addition, this symbiosis influences the distribution and rarity of this medicinally valuable taxon. Specific fungal partners may lead to enhanced seed germination rate and increased efficiency of nutrient exchange between both the partners. -

September - November 2006
Australasian Plant Conservation BULLETIN OF THE AUSTRALIAN NETWORK FOR PLANT CONSERVATION VOLUME 15 NUMBER 2 • SEPTEMBER - NOVEMBER 2006 Role of symbiotic relationships in Australian terrestrial orchid conservation Fungi in agricultural landscapes: implications for eucalypt woodland revegetation Mycorrhizal fungi of Prasophyllum Small mammals as seed dispersers And much much more……… SPECIAL THEME: CONSERVING SYMBIOSES AUSTRALASIAN PLANT CONSERVATION This issue From the Editor: introducing ‘Conserving Symbioses’ 1 Role of symbiotic relationships in Australian terrestrial 2 orchid conservation Endophytic fungi associated with Australian orchids 7 Mycorrhizal fungi of Prasophyllum 10 ANPC Inc. Mission Statement Royal Botanic Gardens Melbourne contributes to Victorian 12 orchid conservation “To promote and improve plant conservation” Below-ground thinking: a review of the 5th International 14 Conference on Mycorrhiza Contributing to Australasian Plant Conservation Fungi in agricultural landscapes: implications for eucalypt 15 woodland revegetation Australasian Plant Conservation is a forum for information exchange for all Small mammals as seed dispersers 19 those involved in plant conservation: FloraBank lives again 21 please use it to share your work with others. Articles, information snippets, The Auckland Botanic Gardens Threatened Native Plant Garden 23 details of new publications or research, and diary dates are welcome. The deadline Regular Features for the December 2006-February 2007 issue is Friday 24 November 2006. Report from New Zealand Plant Conservation Network 25 The December-February issue will be Book Review 26 on the special theme of ‘Conserving Grasslands and Grassy Ecosystems’; Research Roundup 27 however general articles are also very Information Resources and Useful Websites 28 welcome. Please contact Tom May if you are intending to submit an article: ANPC Workshops 29 [email protected]. -

Native Orchid Society of South Australia
NATIVE ORCHID SOCIETY of SOUTH AUSTRALIA JOURNAL NATIVE ORCHID SOCIETY OF SOUTH AUSTRALIA JOURNAL Volume 9, No. 3 April, 1985 Registered by Australia Post Pub. No. SBH 1344 Price 5o¢ PATRON: Mr T.R.N. Lothian CONTENTS: Page 19 Next Meeting PRESIDENT: 19 Hilda Poxon Growers Mr R. Shooter 20 Correspondence Telephone 356 2666 20 New Constitution 20 World First 20 Ira Butler Awards VICE-PRESIDENT: 21 NOSSA Presidents Report - 1985 Mr. K. Western 22 Orchids on Display - March 23 Some Thoughts on the Status of T. x juncifolia. SECRETARY: 24. Help Mr W.K. Harris 25 Orchid Index Telephone 278 2917 25 Diuris longifolia Experiment 27 Orchids of the Warrumbungle Mountains 28 7th Regional Orchid Conference TREASURER: 29 Field Trip to Mt Compass Swamps Mr R.T. Robjohns 30 Show dates for 1985 30 Subscriptions EDITOR: Letizia Gentile NEXT MEETING Telephone 45 6453 When: Tuesday, 23 April, 8.00 p.m. COMMITTEE: Mr R. Bates Where: St Matthews Hall, Bridge Street, Mr. G. Brooks Kensington. Mr. D. Harper Mr. J. Jacobs Subject: Dr. Richard Williams will be speaking to us on the function of Black Hill, and in particular the native orchid section. LIFE MEMBERS: Mr. R. Hargreaves Mr. H. Goldsack Mr R.T. Robjohns Mr. J. Simmons HILDA POXON GROWERS Members who participated in the competition last Postal Address for year please bring their specimen of Hilda Poxon NOSSA: P.O. Box 565 to the April meeting for comparisons to be made. UNLEY. S.A. 5061 20 CORRESPONDENCE I have received a letter dated 12/3/85 from a Mr Rod. -

Action Statement Flora and Fauna Guarantee Act 1988 No
Action Statement Flora and Fauna Guarantee Act 1988 No. 82 Yellow Hyacinth Orchid Dipodium hamiltonianum Description and Distribution The Yellow Hyacinth Orchid (Dipodium In the rain-shadow areas of East Gippsland, two hamiltonianum F.M. Bailey) is a leafless, terrestrial, populations (20 and seven plants respectively) saprophytic orchid with a flower stem up to 95 cm near Wulgulmerang have been recorded, in the tall. It bears 3–25 flowers, each 45 mm across, Alpine National Park (Cobberas-Tingaringy Unit) which are bright yellow to dull yellowish-green and and Snowy River National Park. Rain-shadow bear numerous scattered red or purple spots. The woodland with White Box Eucalyptus albens petals and sepals are widely spread and are not dominates these sites (Clark 1976, Beauglehole reflexed or recurved near the tips. The 20 mm long 1981). There is also an as yet unconfirmed record labellum is narrow and elongate and projects of Yellow Hyacinth Orchid from Suggan Buggan forward; it is pink/purple to white, with a tuft of River (Beauglehole 1981). white hairs extending from the callus to the tip. In the lower rainfall parts of north-eastern Detailed descriptions are given in Jones (1988), Victoria, seven populations have been recorded. Walsh and Entwisle (1994) and Backhouse and Four are in the Beechworth area: one in Jeanes (1995). Beechworth Historic Park (59 plants), two in Mt For most of the year this orchid remains dormant Pilot Multi-Purpose Park (each with two plants), underground as a large, fleshy, branched tuberous and one on private land (1 plant). All are root. -

Endophytic Fungi Associated with Australian Orchids
Endophytic fungi associated with Australian orchids John D. W. Dearnaley and Andrew F. Le Brocque Department of Biological and Physical Sciences, University of Southern Queensland, Toowoomba QLD. Email: [email protected] Fungal endophytes of orchids Australia is rich in orchid flora with over 1000 native species currently recorded. A significant proportion of Australia’s terrestrial orchids are critically endangered, endangered or threatened. Threats to many orchid species include habitat destruction, degradation and fragmentation from increased urbanisation, overgrazing, altered fire regimes and unfortunately, excessive collecting by orchid fanciers. Conservation efforts for Australian orchids include both ex situ and in situ approaches. Ex situ efforts involve the growth of orchid species under horticultural conditions and long term storage of plant and associated fungal material in laboratories and herbaria. In situ approaches include re-establishing plants in the wild and protection of current populations through management initiatives. All orchids depend on fungi for their nutritional needs. As the seeds of orchid are minute and contain very few stored reserves, fungal colonisation is essential for further growth and development following germination (Smith and Read 1997). During plant colonisation fungal hyphae penetrate cells and form elaborate coiled structures known as pelotons (see Figure 1). A peloton is the site of nutrient exchange between plant and fungus and it is via these structures that young orchids receive sugars and inorganic substances (eg. P, N) necessary for further growth. Because the fungi grow within plant cells they are called endophytes. Mature photosynthetic orchids remain colonised by fungi and supply their endophytes with sugars but continue to receive inorganic nutrients. -

Ectomycorrhizal Inocybe Species Associate with the Mycoheterotrophic Orchid Epipogium Aphyllum but Not Its Asexual Propagules
Annals of Botany 104: 595–610, 2009 doi:10.1093/aob/mcn269, available online at www.aob.oxfordjournals.org Ectomycorrhizal Inocybe species associate with the mycoheterotrophic orchid Epipogium aphyllum but not its asexual propagules Melanie Roy1,†, Takahiro Yagame2, Masahide Yamato3, Koji Iwase4, Christine Heinz5, Antonella Faccio6, Paola Bonfante6 and Marc-Andre Selosse1,†,* 1Centre d’Ecologie Fonctionnelle et Evolutive (CNRS, UMR 5175), Equipe Interactions Biotiques, 1919 Route de Mende, 34293 Montpellier ce´dex 5, France, 2Orchid Museum Takamori, 512-73 Izuhara, Shimoina, Nagano 399-3107, Japan, 3Environment Department, The General Environmental Technos Co., Ltd, 1-3-5 Azuchimachi Chuo-ku, Osaka 541-0052, Japan, 4Fungus/ Mushroom Resource and Research Center, Faculty of Agriculture, Tottori University, 4-101 Koyama-minami, Tottori 680-8553, Japan, 5Universite´ Montpellier 2, UMR AMAP Botanique et bioinformatique de l’Architecture des Plantes, 34000 Montpellier, France and 6Dipartimento di Biologia Vegetale dell’Universita`, Istituto per la Protezione delle Piante – CNR, Viale Mattioli 25, 10125 Torino, Italy Received: 1 June 2008 Returned for revision: 22 September 2008 Accepted: 25 November 2008 Published electronically: 19 January 2009 † Background and Aims Epipogium aphyllum is a Eurasian achlorophyllous, mycoheterotrophic forest orchid. Due to its rarity, it is often protected, and its biology is poorly known. The identity and pattern of colonization of fungal associates providing carbon to this orchid have not been studied previously. † Methods Using samples from 34 individuals from 18 populations in Japan, Russia and France, the following were investigated: (a) colonization patterns of fungal associates of E. aphyllum by microscopy; (b) their identity by PCR amplification of nuclear ribosomal ITS carried out on rhizome fragments and hyphal pelotons. -

Dubbo Region Flora List 2012
Flora List of the Dubbo Area and Central Western Slopes Harlequin Mistletoe Lysiana exocarpi subsp. tenuis Drilliwarrina State Conservation Area Janice Hosking for the Dubbo Field Naturalist and Conservation Society Inc Version: June 2012 www.dubbofieldnats.org.au Flora List of the Dubbo Area and Central Western Slopes Janice Hosking for Dubbo Field Nats This list of approximately 1,300 plant species was prepared by Janice Hosking for the Dubbo Field Naturalist & Conservation Society Inc. Many thanks to Steve Lewer and Chris McRae who spent many hours checking and adding to this list. Cover photo: Anne McAlpine, A map of the area subject to this list is provided below. Data Sources: This list has been compiled from the following information: A Flora of the Dubbo District 25 Miles radius around the city (c. 1950s) compiled by George Althofer, assisted by Andy Graham. Gilgandra Native Flora Reserve Plant List Goonoo State Forest Forestry Commission list, supplemented by Mr. P. Althofer. List No.1 (c 1950s) Goonoo State Forest Dubbo Management Area list of Plants List No.2 The Flora of Mt. Arthur Reserve, Wellington NSW A small list for Goonoo State Forest. Author and date unknown Flora List from Cashells Dam Area, Goonoo State Forest (now CCA) – compiled by Steve Lewer (NSW OEH) Oasis Reserve Plant List (Southwest of Dubbo) – compiled by Robert Gibson (NSW OEH) NSW DECCW Wildlife atlas List 2010,Y.E.T.I. List 2010 PlantNet (NSW Botanic Gardens Records) Various species lists for Dubbo District rural properties – compiled by Steve Lewer (NSW OEH) * Denotes an exotic species ** Now considered to be either locally extinct or possibly a misidentification. -
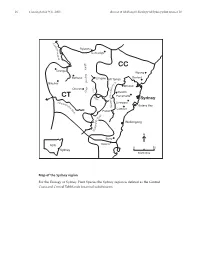
Part 10 ESP Intro
16 Cunninghamia 9(1): 2005 Benson & McDougall, Ecology of Sydney plant species 10 M a c q u Rylstone a r i e Coricudgy R i v e r e g n CC a Orange R Wyong g n i Gosford Bathurst d i Lithgow v Mt Tomah i Blayney D R. y r Windsor C t u a o b Oberon s e x r e s G k Penrith w a R Parramatta CT H i ve – Sydney r n a Abe e Liverpool rcro p m e b Botany Bay ie N R Camden iv Picton er er iv R y l l i Wollongong d n o l l o W N Berry NSW Nowra 050 Sydney kilometres Map of the Sydney region For the Ecology of Sydney Plant Species the Sydney region is defined as the Central Coast and Central Tablelands botanical subdivisions. Cunninghamia 9(1): 2005 Benson & McDougall, Ecology of Sydney plant species 10 17 Ecology of Sydney plant species Part 10 Monocotyledon families Lemnaceae to Zosteraceae Doug Benson and Lyn McDougall Royal Botanic Gardens and Domain Trust, Sydney, AUSTRALIA 2000. Email: [email protected] Abstract: Ecological data in tabular form are provided on 668 plant species of the families Lemnaceae to Zosteraceae, 505 native and 163 exotics, occurring in the Sydney region, defined by the Central Coast and Central Tablelands botanical subdivisions of New South Wales (approximately bounded by Lake Macquarie, Orange, Crookwell and Nowra). Relevant Local Government Areas are Auburn, Ashfield, Bankstown, Bathurst, Baulkham Hills, Blacktown, Blayney, Blue Mountains, Botany, Burwood, Cabonne, Camden, Campbelltown, Canada Bay, Canterbury, Cessnock, Crookwell, Evans, Fairfield, Greater Lithgow, Gosford, Hawkesbury, Holroyd, Hornsby, Hunters Hill, Hurstville, Kiama, Kogarah, Ku-ring-gai, Lake Macquarie, Lane Cove, Leichhardt, Liverpool, Manly, Marrickville, Mosman, Mulwaree, North Sydney, Oberon, Orange, Parramatta, Penrith, Pittwater, Randwick, Rockdale, Ryde, Rylstone, Shellharbour, Shoalhaven, Singleton, South Sydney, Strathfield, Sutherland, Sydney City, Warringah, Waverley, Willoughby, Wingecarribee, Wollondilly, Wollongong, Woollahra and Wyong.