Development and External Validation of a COVID-19 Mortality Risk Prediction Algorithm: a Multicentre Retrospective Cohort Study
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Present Status, Driving Forces and Pattern Optimization of Territory in Hubei Province, China Tingke Wu, Man Yuan
World Academy of Science, Engineering and Technology International Journal of Environmental and Ecological Engineering Vol:13, No:5, 2019 Present Status, Driving Forces and Pattern Optimization of Territory in Hubei Province, China Tingke Wu, Man Yuan market failure [4]. In fact, spatial planning system of China is Abstract—“National Territorial Planning (2016-2030)” was not perfect. It is a crucial problem that land resources have been issued by the State Council of China in 2017. As an important unordered and decentralized developed and overexploited so initiative of putting it into effect, territorial planning at provincial level that ecological space and agricultural space are seriously makes overall arrangement of territorial development, resources and squeezed. In this regard, territorial planning makes crucial environment protection, comprehensive renovation and security system construction. Hubei province, as the pivot of the “Rise of attempt to realize the "Multi-Plan Integration" mode and Central China” national strategy, is now confronted with great contributes to spatial planning system reform. It is also opportunities and challenges in territorial development, protection, conducive to improving land use regulation and enhancing and renovation. Territorial spatial pattern experiences long time territorial spatial governance ability. evolution, influenced by multiple internal and external driving forces. Territorial spatial pattern is the result of land use conversion It is not clear what are the main causes of its formation and what are for a long period. Land use change, as the significant effective ways of optimizing it. By analyzing land use data in 2016, this paper reveals present status of territory in Hubei. Combined with manifestation of human activities’ impact on natural economic and social data and construction information, driving forces ecosystems, has always been a specific field of global climate of territorial spatial pattern are then analyzed. -

3. Integrated Pest Management Plan
Appendix 4 Public Disclosure Authorized Public Disclosure Authorized World Bank Financed Project Hubei Safe, Sustainable, Smart Agricultural Project Pest Management Framework Public Disclosure Authorized Public Disclosure Authorized Submitted by: Foreign Cooperation Office of Hubei Department of Agriculture and Rural Affairs Prepared by: Central-Southern Safety&Environment Technology Institute Co., LTD November 2019 Table of Contents 1. Project Overview .....................................................................................................1 2. Project Backgrounds ...............................................................................................3 2.1 Project Objectives .............................................................................................3 2.2 Pest Problems of Project Crops ........................................................................4 2.3 Current Situation of Chemical Pesticide Use ...................................................5 2.4 Crop Pest Management and Problems ..............................................................5 2.5 Possible Environmental Impacts and Risks after Project Implementation .......7 2.6 Evaluation of Existing Policies and Systems ....................................................9 2.7 Institutional Framework for Pest Management .............................................. 11 3. Integrated Pest Management Plan .........................................................................18 3.1 Objectives and Priorities .................................................................................18 -

Hubei Safe, Sustainable and Smart Agriculture Project (Hereinafter, the “Project”)
World Bank-financed Project Public Disclosure Authorized Hubei Safe, Sustainable and Smart Public Disclosure Authorized Agriculture Project Resettlement Policy Framework Public Disclosure Authorized Public Disclosure Authorized Hubei Project Management Office (PMO) October 2019 Foreword I. Purpose of preparing this RPF 1 The RPF is prepared in accordance with the applicable laws of the People’s Republic of China and local regulations and a series of provisions in the Bank Operational Policy OP4.12 on Involuntary Resettlement for the purpose of “developing an action plan for resettlement and restoration for the people affected by the project, so that they benefit from the project, their living standard is improved or at least restored after the completion of the project”. II. Definitions of terms Displaced persons 2 Based on the criteria for eligibility for compensation, “Displaced Persons” may be classified in one of the following three groups: a) those who have formal legal rights to land (including customary and traditional rights recognized under the laws of the country); b) those who do not have formal legal rights to land at the time the census begins but have a claim to such land or assets-provided that such claims are recognized under the laws of the country or become recognized through a process identified in the RPF; and c) those who have no recognizable legal right or claim to the land they are occupying. 3 Persons covered under paragraphs 2(a) and (b) are provided compensation for the land they lose, and other assistance. Persons covered under paragraph 2(c) are provided resettlement assistance in lieu of compensation for the land they occupy, and other assistance, as necessary, to achieve the objective set out in this policy, if they occupy the project area prior to a cut-off date1 established by the borrower and acceptable to the World Bank. -

Hubei Xiangyang Integrated Sustainable Transportation and Logistics Planning and Strategic Study
Technical Assistance Consultant’s Report Project Number: 51029-001 June 2020 People’s Republic of China: Hubei Xiangyang Integrated Sustainable Transportation and Logistics Planning and Strategic Study Prepared by: David Lupton & Associates Ltd. For the Asian Development Bank and Xiangyang Municipal Transport Bureau This consultant’s report does not necessarily reflect the views of ADB or the Government concerned, and ADB and the Government cannot be held liable for its contents. TA-9547 PRC: Hubei Xiangyang Comprehensive Transportation and Logistics Planning and Strategic Study- 01 (51029-001) Supplementary Report June 2020 Prepared for: the Asian Development Bank and Xiangyang Municipal Transport Bureau David Lupton & Associates Ltd. TA-9547 PRC: Hubei Xiangyang Comprehensive Transportation and Logistics Planning and Strategic Study- 01 (51029-001) CURRENCY EQUIVALENT (as of 10 January 2019) Currency unit: USD USD1=7.08 CNY 1 CNY=USD 0.14 WEIGHTS AND MEASURES km2 – square kilometre m3 cubic meter mu: 15 mu=1 hectare NOTES (i) In this Report, “$” refers to US Dollars (ii) The fiscal year (FY) of the Government and its agencies ends on 31 December. FY before a calendar year denotes the year in which the fiscal year ends, e.g., FY2018 ends on 31 December 2018. (iii) At the 17 January 2019 meeting between the client the XMTB, ADB and the consultant, the client requested that the title for the Project be changed to “Hubei Xiangyang Comprehensive Transportation and Logistics Planning and Strategic Study” in order to reflect the new Han River Economic Development Plan Consultants Quality Assurance Protocol Prepared By Robert L. Wallack, David J. -

Independent Technical Report of Xiangyang Future Bright Mining's
THIS DOCUMENT IS IN DRAFT FORM, INCOMPLETE AND SUBJECT TO CHANGE AND THE INFORMATION MUST BE READ IN CONJUNCTION WITH THE SECTION HEADED “WARNING” ON THE COVER OF THIS DOCUMENT. APPENDIX IV INDEPENDENT TECHNICAL REPORT Independent Technical Report of Xiangyang Future Bright Mining’s Yiduoyan Marble Project Report Prepared for Future Bright Mining Holdings Limited Report Prepared by SRK Consulting (Hong Kong) Limited XFB002 [REDACTED] – IV-1 – THIS DOCUMENT IS IN DRAFT FORM, INCOMPLETE AND SUBJECT TO CHANGE AND THE INFORMATION MUST BE READ IN CONJUNCTION WITH THE SECTION HEADED “WARNING” ON THE COVER OF THIS DOCUMENT. APPENDIX IV INDEPENDENT TECHNICAL REPORT Independent Technical Report of Xiangyang Future Bright Mining’s Yiduoyan Marble Project Future Bright Mining Holdings Limited R16/F, Guangdong Finance Building, 88 Connaught Road West, Hong Kong SRK Consulting (Hong Kong) Limited Suite A1, 11/F, One Capital Place, 18 Luard Road, Wanchai, Hong Kong e-mail: [email protected] website: srk.com.hk Tel: +852 2520 2522 Fax: +852 2520 0003 SRK Project Number XFB002 [REDACTED] Compiled by Peer Reviewed by Helen Ray Anthony Stepcich Associate, Industrial Minerals Principal Consultant, Project Evaluations Authors: Gavin Chan; Helen Ray; Sue Border; Jinhui Liu; Gerard Guo; Xue Nan – IV-2 – THIS DOCUMENT IS IN DRAFT FORM, INCOMPLETE AND SUBJECT TO CHANGE AND THE INFORMATION MUST BE READ IN CONJUNCTION WITH THE SECTION HEADED “WARNING” ON THE COVER OF THIS DOCUMENT. APPENDIX IV INDEPENDENT TECHNICAL REPORT EXECUTIVE SUMMARY SRK Consulting (Hong Kong) Limited (“SRK”) has been commissioned by Future Bright Mining Holdings Limited (“Future Bright” or the “Company”) to prepare an Independent Technical Report (“ITR” or “Report”) on its Yiduoyan Marble Project (the “Project”), located in Xiaoyan Township, Nanzhang County, Xiangyang, Hubei Province of Peoples’ Republic of China. -

Research on the Ecology Spatial Pattern of Basic Farmland Guided by the Concept of “Negative Planning” X
Bulgarian Chemical Communications, Volume 49, Special Edition K2, (pp. 144 – 151) 2017 Research on the ecology spatial pattern of basic farmland guided by the concept of “negative planning” X. Lan*, Z. Q. Wang, C. Wei, L. G. Zhang School of Public Management, China University of Geosciences (CUG), No.388 Lumo Road, Wuhan 430074, China Received August 15, 2017, Revised November 15, 2017 After the 19th National Congress of the Communist Party of China had been launched successfully, the Chinese government showed confidence by improving the ecological environment of the whole country in changing the traditional planning method which considers the social and economic benefits rather than the ecological one. In this paper, the simulation experiment tries to delimit an area of ecological infrastructure with the guidance of the “security pattern” and “negative planning” concept which has a more favorable index of ecological environment than the traditional way in the spatial pattern planning of basic farmland. The results show that the new planning provides more high-quality and less fragmentary cultivated land than the traditional one, and the cultivated land is more adaptable in biodiversity and producing activity, which is in line with the development of environmental policies of the Chinese government and the global sustainable development. Keywords: Negative planning, Security pattern of ecological infrastructure, Basic farmland protection, Spatial pattern, Baokang county INTRODUCTION division [3-6]. In China, some scholars have proposed that when protecting arable land, we As a core part of Chinese “keep farmland in 1.8 should pay more attention to its multifunctionality, billion mu (approximately 120 million hectares)” specify the reasonable direction of usage and carry policy, the protection of basic farmland should be out the practical plans for usage, enhancing the attended to with great importance, whether in laws mobility of protecting the cultivated land [7-9]. -

Harmonious Development Between Socio-Economy and River-Lake Water Systems in Xiangyang City, China
water Article Harmonious Development between Socio-Economy and River-Lake Water Systems in Xiangyang City, China Qiting Zuo 1,2, Zengliang Luo 1 and Xiangyi Ding 3,* 1 School of Water Conservancy & Environment, Zhengzhou University, Zhengzhou 450001, China; [email protected] (Q.Z.); [email protected] (Z.L.) 2 Center for Water Science Research, Zhengzhou University, Zhengzhou 450001, China 3 Department of Water Resources, China Institute of Water Resources and Hydropower Reasearch, Beijing 100038, China * Correspondence: [email protected]; Tel.: +86-10-6878-1373 Academic Editor: Karl-Erich Lindenschmidt Received: 18 September 2016; Accepted: 31 October 2016; Published: 4 November 2016 Abstract: River-lake water systems (RLS) are important carriers for matter transformation and energy transmission. Influenced by accelerated social and economic development, the structural, functional, and environmental states of RLS have been seriously damaged. It is an important problem for human beings to coordinate the contradiction between socio-economic development and the protection of RLS. In order to quantitatively study the harmonious relationship between socio-economic development and the state of RLS, the harmony theory method was used to analyze the degree of harmonious development between socio-economy and RLS in this study taking Xiangyang City as an example, and formulating corresponding harmonious optimization schemes. The results indicate that: (1) the state of RLS had a relatively small change during 2009–2014, and its spatial -

P020201230847455570044.Pdf
/ The Speech at the Provincial Water Conservancy Financial Work Video Conference Wang Zhongfa (1) Summary Speech at the Provincial Water Conservancy Financial Work Video Conference Feng Zhongkai (5) Work Report at the Provincial Water Conservancy Financial Work Video Conference Shao Yuanliang(7) Play the Role of Financial Functions to Support Water Conservancy Reform and Development Wang Yi (10) The Speech at the Provincial Water Conservancy Financial Work Video Conference Li Jie (12) The Speech at the Provincial Water Conservancy Financial Work Video Conference Li Shunzhou(13) Explore the New Mode of Water Conservancy Capital Regulation to Guarantee of Water Conservancy Reform and Development in Xiangyang City Zou Hongcheng (14) Strengthen the Department to Budget Regulate Fund Supervision in Jinzhou City Hao Yongyao(16) Promoting Reform Practice on the Water Conservancy Separation of Custody in Yunxi County Xu Qingdong(18) Innovation Mechanism,Actively Promote Agricultural Water Price Reform in Yuanan County Huang Guilin (20) Work Practices on the Reform of Small Water Conservancy Facilities Management System in Suixian County Shen Yichun(21) Thinking on Improving the Management Level of Hubei Hydrology Financial Management Liu Zuoyin (22) Exploration on the Reform of Small Water Conservancy Facilities Management System in Yiling District Yichang City Water Bureau of Yiling District Yichang City (24) Exploration on the Reform of Small Water Conservancy Facilities Management System in Lao Hekou City Water Bureau of Lao Hekou city Hubei -

Explanations on Revision on Resettlement Action Plan
RPII0U Volume 1 The World Bank Financed Project Xiao-xiangExpressway, Hubei,China Public Disclosure Authorized RESETTLEMENT ACTION PLAN Public Disclosure Authorized Public Disclosure Authorized Hubei Province Xiao-xiang Expressway Resettlement Office Public Disclosure Authorized January 2002 Wuhan, China FIL COPY Explanations on Revision on Resettlement Action Plan Revision on Compensation Standards 1. Standard of expense for water supply, power connection and traffic as well as leveling the land for reconstruction is altered from "5 Yuan/Mr2 " to "375 Yuan per family whose resettlement house area is not larger than 100m2"/"750 Yuan per family whose resettlement house area is larger than 100m2". Compensation standard for family transit cost/ delay of their work and other subsidy are 150 Yuan per family. Therefore, cost of this item is increased from 962443 Yuan to 1186050 Yuan. Increased amount is 223607 Yuan. (This revision should be affirmed by higher institutions). 2. Compensation standard on brick & concrete structure is lifted from 21OYuan/m 2 to 22OYuan/m 2, which is determined by Mr. Zhang, Xuefeng (Vise Director of HPCD) and Mr. Liu, Zefu. Therefore, cost of this item is increased from 13588008 Yuan to 14235056 Yuan. Increased amount is 647048 Yuan. 3. Capital flow direction in the Capital Flowchart is consulted between Mr. Zhang, Xuefeng and Mr. Liu, Zefu. It is determined that account for land compensation should be opened in local county banks by county-level resettlement institutions. Fund application plan should be put forward by village administrative committee(VAC) and be approved by relevant county/town institutions before drawing from banks who can otherwise refuse the payment. -
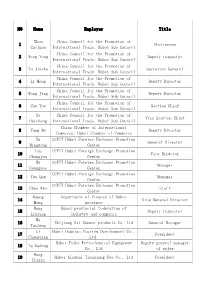
List of the Delegation of Hubei Province
NO Name Employer Title Zhou China Council for the Promotion of 1 Chairwoman Caijuan International Trade, Hubei Sub-Council China Council for the Promotion of 2 Wang Yong Deputy inspector International Trade, Hubei Sub-Council China Council for the Promotion of 3 Yu Jiazhu Secretary General International Trade, Hubei Sub-Council China Council for the Promotion of 4 Li Meng Deputy Director International Trade, Hubei Sub-Council China Council for the Promotion of 5 Wang Jing Deputy Director International Trade, Hubei Sub-Council China Council for the Promotion of 6 Cao Yue Section Chief International Trade, Hubei Sub-Council Xu China Council for the Promotion of 7 Vice Section Chief Huichang International Trade, Hubei Sub-Council China Chamber of International 8 Yang Bo Deputy Director Commerce, Hubei Chamber of Commerce Xu CCPIT Hubei Foreign Exchange Promotion 9 General Director Mingsong Center Lyu CCPIT Hubei Foreign Exchange Promotion 10 Vice Director Chongjun Center Hu CCPIT Hubei Foreign Exchange Promotion 11 Manager Guangyao Center CCPIT Hubei Foreign Exchange Promotion 12 Yue Qun Manager Center CCPIT Hubei Foreign Exchange Promotion 13 Chen Wen Staff Center Huang Department of Finance of Hubei 14 Vice General Director Ming province Wang Hubei provincial federation of 15 Deputy inspector Lixiang industry and commerce Wu 16 Zhijiamg Exi Cement products Co.,Ltd General Manager Taifeng Li Hubei Houniao Tourism Development Co., 17 President Changqing Ltd. Hubei Taihe Petrochemical Equipment Deputy general manager 18 Ye Kedong Co., Ltd -

Minimum Wage Standards in China June 28, 2018
Minimum Wage Standards in China June 28, 2018 Contents Heilongjiang .................................................................................................................................................. 3 Jilin ................................................................................................................................................................ 3 Liaoning ........................................................................................................................................................ 4 Inner Mongolia Autonomous Region ........................................................................................................... 7 Beijing ......................................................................................................................................................... 10 Hebei ........................................................................................................................................................... 11 Henan .......................................................................................................................................................... 13 Shandong .................................................................................................................................................... 14 Shanxi ......................................................................................................................................................... 16 Shaanxi ....................................................................................................................................................... -

A12 List of China's City Gas Franchising Zones
附录 A12: 中国城市管道燃气特许经营区收录名单 Appendix A03: List of China's City Gas Franchising Zones • 1 Appendix A12: List of China's City Gas Franchising Zones 附录 A12:中国城市管道燃气特许经营区收录名单 No. of Projects / 项目数:3,404 Statistics Update Date / 统计截止时间:2017.9 Source / 来源:http://www.chinagasmap.com Natural gas project investment in China was relatively simple and easy just 10 CNG)、控股投资者(上级管理机构)和一线运营单位的当前主官经理、公司企业 years ago because of the brand new downstream market. It differs a lot since 所有制类型和联系方式。 then: LNG plants enjoyed seller market before, while a LNG plant investor today will find himself soon fighting with over 300 LNG plants for buyers; West East 这套名录的作用 Gas Pipeline 1 enjoyed virgin markets alongside its paving route in 2002, while today's Xin-Zhe-Yue Pipeline Network investor has to plan its route within territory 1. 在基础数据收集验证层面为您的专业信息团队节省 2,500 小时之工作量; of a couple of competing pipelines; In the past, city gas investors could choose to 2. 使城市燃气项目投资者了解当前特许区域最新分布、其他燃气公司的控股势力范 sign golden areas with best sales potential and easy access to PNG supply, while 围;结合中国 LNG 项目名录和中国 CNG 项目名录时,投资者更易于选择新项 today's investors have to turn their sights to areas where sales potential is limited 目区域或谋划收购对象; ...Obviously, today's investors have to consider more to ensure right decision 3. 使 LNG 和 LNG 生产商掌握采购商的最新布局,提前为充分市场竞争做准备; making in a much complicated gas market. China Natural Gas Map's associated 4. 便于 L/CNG 加气站投资者了解市场进入壁垒,并在此基础上谨慎规划选址; project directories provide readers a fundamental analysis tool to make their 5. 结合中国天然气管道名录时,长输管线项目的投资者可根据竞争性供气管道当前 decisions. With a completed idea about venders, buyers and competitive projects, 格局和下游用户的分布,对管道路线和分输口建立初步规划框架。 analyst would be able to shape a better market model when planning a new investment or marketing program.