Sanger Sequencing – a Hands-On Simulation Background And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Bacteriophage T7 DNA Polymerase – Sequenase
Bacteriophage T7 DNA polymerase – sequenase The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Zhu, Bin. 2014. “Bacteriophage T7 DNA polymerase – sequenase.” Frontiers in Microbiology 5 (1): 181. doi:10.3389/fmicb.2014.00181. http://dx.doi.org/10.3389/fmicb.2014.00181. Published Version doi:10.3389/fmicb.2014.00181 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:12407012 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA REVIEW ARTICLE published: 16 April 2014 doi: 10.3389/fmicb.2014.00181 BacteriophageT7 DNA polymerase – sequenase Bin Zhu* Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA, USA Edited by: An ideal DNA polymerase for chain-terminating DNA sequencing should possess the Andrew F.Gardner, New England following features: (1) incorporate dideoxy- and other modified nucleotides at an efficiency Biolabs, USA similar to that of the cognate deoxynucleotides; (2) high processivity; (3) high fidelity in Reviewed by: the absence of proofreading/exonuclease activity; and (4) production of clear and uniform Kirk Matthew Schnorr, Novozymes A/S, Denmark signals for detection. The DNA polymerase encoded by bacteriophage T7 is naturally Samir Hamdan, King Abdullah endowed with or can be engineered to have all these characteristics. The chemically or University of Science and Technology, genetically modified enzyme (Sequenase) expedited significantly the development of DNA Saudi Arabia sequencing technology. -

Sequencing.Pdf
Sequencing. Biological sciences has a long and storied history of natural science – identifying, cataloging, and classifying organisms in different environments. Every aspect of biological sciences is being revolutionized by two dominant technologies: cheap and ubiquitous access to DNA sequencing, and cheap and ubiquitous access to high performance computing. Molecular biology was forever altered by the introduction of the polymerase chain reaction (PCR), and that invention, more than any other, has enabled the sequencing revolution. Taxonomy, one of the oldest endeavors of the biologist, is changing from char- acterizing organisms based on morphology to characterizing organisms based on DNA sequences. Revisionist taxonomic approaches are being introduced throughout the tree of life, and no branch on the tree can avoid the sequenc- ing machine. For example, microbial and viral taxonomy are being completely rewritten because of the analysis of DNA sequences (Rohwer & Edwards, 2002; Thompson et al., 2015). All aspects of ecology are being affected by the intro- duction of DNA sequencing: for example, in the marine environment the ecology of everything from the largest (Cammen et al., 2016) to the smallest (Chow & Suttle, 2015; Haggerty & Dinsdale, 2017) organisms is being revised because of sequence analysis. Evolutionary theories about plants and animals are being upended because of the introduction of genome skimming techniques and deep sequencing (Hollingsworth et al., 2016). Sanger Sequencing Two-time nobel Laureate Dr. Fred Sanger developed this sequencing technol- ogy in 1977. The approach uses terminators, dideoxynucleotide triphosphates (ddNTPs), that terminate DNA extension during replication and do not allow the strand to be synthesized further. Thus, every time a ddNTP is added to the growing DNA strand, synthesis stops and the fragement can not be made any longer. -

DNA Sequencing with Thermus Aquaticus DNA Polymerase
Proc. Natl. Acad. Sci. USA Vol. 85, pp. 9436-9440, December 1988 Biochemistry DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA (thermophilic DNA polymerase/chain-termination/processivity/automation) MICHAEL A. INNIS*, KENNETH B. MYAMBO, DAVID H. GELFAND, AND MARY ANN D. BROW Department of Microbial Genetics, Cetus Corporation, 1400 Fifty-Third Street, Emeryville, CA 94608 Communicated by Hamilton 0. Smith, September 8, 1988 (receivedfor review August 15, 1988) ABSTRACT The highly thermostable DNA polymerase properties of Taq DNA polymerase that pertain to its advan- from Thermus aquaticus (Taq) is ideal for both manual and tages for DNA sequencing and its fidelity in PCR. automated DNA sequencing because it is fast, highly proces- sive, has little or no 3'-exonuclease activity, and is active over a broad range of temperatures. Sequencing protocols are MATERIALS presented that produce readable extension products >1000 Enzymes. Polynucleotide kinase from T4-infected Esche- bases having uniform band intensities. A combination of high richia coli cells was purchased from Pharmacia. Taq DNA reaction temperatures and the base analog 7-deaza-2'- polymerase, a single subunit enzyme with relative molecular deoxyguanosine was used to sequence through G+C-rich DNA mass of 94 kDa (specific activity, 200,000 units/mg; 1 unit and to resolve gel compressions. We modified the polymerase corresponds to 10 nmol of product synthesized in 30 min with chain reaction (PCR) conditions for direct DNA sequencing of activated salmon sperm DNA), was purified from Thermus asymmetric PCR products without intermediate purification aquaticus, strain YT-1 (ATCC no. -

DNA Sequencing
DNA Sequencing: Sanger Method (Dideoxynucleotide chain termination) Sanger sequencing is a DNA sequencing method in which target DNA is denatured and annealed to an oligonucleotide primer, which is then extended by DNA polymerase using a mixture of deoxynucleotide triphosphates (normal dNTPs) and chain-terminating dideoxynucleotide triphosphates (ddNTPs). ddNTPs lack the 3’ OH group to which the next dNTP of the growing DNA chain is added. Without the 3’ OH, no more nucleotides can be added, and DNA polymerase falls off. The resulting newly synthesized DNA chains will be a mixture of lengths, depending on how long the chain was when a ddNTP was randomly incorporated. Manual DNA sequencing example: • First, anneal the primer to the DNA template (must be single stranded): 5’ -GAATGTCCTTTCTCTAAG 3'-GGAGACTTACAGGAAAGAGATTCAGGATTCAGGAGGCCTACCATGAAGATCAAG-5' • Then split the sample into four aliquots including the following nucleotides: "G" tube: All four dNTPs, one of which is radiolabeled, plus ddGTP (low concentration) "A" tube: All four dNTPs, one of which is radiolabeled, plus ddATP "T" tube: All four dNTPs, one of which is radiolabeled, plus ddTTP "C" tube: All four dNTPs, one of which is radiolabeled, plus ddCTP • When a DNA polymerase (e.g. Klenow fragment) is added to the tubes, the synthetic reaction proceeds until, by chance, a dideoxynucleotide is incorporated instead of a deoxynucleotide. This is a "chain termination" event, because there is a 3' H instead of a 3' OH group. Since the synthesized DNA is labeled (classically with 35S-dATP), the products can be detected and distinguished from the template. Note that the higher the concentration of the ddNTP in the reaction, the shorter the products will be, hence, you will get sequence CLOSER to your primer. -

Sanger Sequencing 14
Molecular Biology and History of DNA Sequencing 02-223 Sept. 9 2014 History of DNA Thomas Morgan first James Watson and described Francis Crick proposed Gregor Mendel first linkage and that DNA is a double described patterns of recombination strand with a double inheritance helical structure 1866 1869 1911 1950 1953 Fredrich Edwin Chargaff Miescher first discovered that A and isolated DNA T, and G and C have equal amounts http://www.nature.com/scitable/content/dna-is-a-double-helix-24263 History of DNA Frederick Sanger Arthur Kornberg Hamilton Smith developed dideoxy replicated DNA in- discovered DNA Commercial DNA sequencing ~100 vitro using DNA restriction automated DNA bases/reaction polymerase enzymes synthesizer PCR developed ~1000 bases/ by Kary Mullis reaction 1957 1961 1970 1971 1977 1983 1986 1996 First genome sequenced using Leroy Hood Marshall in-vitro replication by Ray Wu, developed Nirenberg A.D. Kaiser, and Ellen Taylor . automated elucidated the Phage λ, ~5000 nt took over 3 sequencing codons years DNA Polymerase h"p://www.virology.ws/2009/05/10/the-error-prone-ways-of-rna-synthesis/ Even with proofreading, mistakes made every 107-109 Bases 6 Billion Bases in human genome! h"p://www.virology.ws/2009/05/10/the-error-prone-ways-of-rna-synthesis/ Molecular Biology of the Cell. 4th edition. Alberts B, Johnson A, Lewis J, et al. New York: Garland Science; 2002. PCR • Polymerase Chain ReacJon • Invented in 1983 • DNA polymerase from Thermus aqua+cus • 2.2x105 error rate Polymerase Chain Reaction (PCR) overview buffer, ssDNA primers, -

A Beginner's Guide to Next Generation Sequencing
A BEGINNER’S GUIDE TO NEXT GENERATION SEQUENCING youseq.com Next Generation Sequencing Let’s keep things simple. The world of Next Generation Sequencing (NGS) can seem complex and intimidating. It need not be. Let’s start by reminding ourselves what its useful for and why we use it. All of life is coded in it’s DNA. A remarkably simple code of four molecules that act as a blue print to define the proteins that we and all of the organisms we share our planet with are made of. Reading this code is one of the most astonishing achievements that the human species has ever and will ever accomplish. Reading this code helps us to understand how we are made, how we are all related, how errors or mutations in our DNA cause disease and how we may respond best to medicines. It holds the promise to revolutionise healthcare and has already begun to do so. The first human genome “read” was competed in 2001. It took 10 years and the best part of $2.7bn. It was achieved by DNA sequencing. A method by which the sequence of the DNA is read painstakingly in small fragments and then reassembled to create a complete sequence. Sanger Sequencing as it is known, was the method used to achieve the first publication of the first human genome. Next Generation Sequencing is a phrase used to describe a range of technologies that speed up and reduce the cost of DNA sequencing vs the traditional Sanger sequencing. What are all these different Next Generation Sequencing Technologies? Well there are quite a few of them. -

Discovery and Characterization Of
Discovery and characterization of microsatellites for the solitary bee Colletes inaequalis using Sanger and 454 pyrosequencing Margarita López-Uribe, Christine Santiago, Steve Bogdanowicz, Bryan Danforth To cite this version: Margarita López-Uribe, Christine Santiago, Steve Bogdanowicz, Bryan Danforth. Discovery and characterization of microsatellites for the solitary bee Colletes inaequalis using Sanger and 454 py- rosequencing. Apidologie, Springer Verlag, 2013, 44 (2), pp.163-172. 10.1007/s13592-012-0168-3. hal-01201284 HAL Id: hal-01201284 https://hal.archives-ouvertes.fr/hal-01201284 Submitted on 17 Sep 2015 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Apidologie (2013) 44:163–172 Original article * INRA, DIB and Springer-Verlag France, 2012 DOI: 10.1007/s13592-012-0168-3 Discovery and characterization of microsatellites for the solitary bee Colletes inaequalis using Sanger and 454 pyrosequencing 1 1 2 Margarita M. LÓPEZ-URIBE , Christine K. SANTIAGO , Steve M. BOGDANOWICZ , 1 Bryan N. DANFORTH 1Department of Entomology, Cornell University, Ithaca, NY 14853, USA 2Evolutionary Genetics Core Facilities, Department of Ecology and Evolutionary Biology, Cornell University, Ithaca, NY 14853, USA Received 5 June 2012 – Revised 8 August 2012 – Accepted 18 September 2012 Abstract – The recent implementation of next-generation sequencing for the discovery of microsatellite markers has made this technology the most effective method for generating genetic markers in non-model organisms. -

12.2% 122000 135M Top 1% 154 4800
View metadata, citation and similar papers at core.ac.uk brought to you by CORE We are IntechOpen, provided by IntechOpen the world’s leading publisher of Open Access books Built by scientists, for scientists 4,800 122,000 135M Open access books available International authors and editors Downloads Our authors are among the 154 TOP 1% 12.2% Countries delivered to most cited scientists Contributors from top 500 universities Selection of our books indexed in the Book Citation Index in Web of Science™ Core Collection (BKCI) Interested in publishing with us? Contact [email protected] Numbers displayed above are based on latest data collected. For more information visit www.intechopen.com 1 DNA Representation Bharti Rajendra Kumar B.T. Kumaon Institute of Technology, Dwarahat,Almora, Uttarakhand, India 1. Introduction The term DNA sequencing refers to methods for determining the order of the nucleotides bases adenine,guanine,cytosine and thymine in a molecule of DNA. The first DNA sequence were obtained by academic researchers,using laboratories methods based on 2- dimensional chromatography in the early 1970s. By the development of dye based sequencing method with automated analysis,DNA sequencing has become easier and faster. The knowledge of DNA sequences of genes and other parts of the genome of organisms has become indispensable for basic research studying biological processes, as well as in applied fields such as diagnostic or forensic research. DNA is the information store that ultimately dictates the structure of every gene product, delineates every part of the organisms. The order of the bases along DNA contains the complete set of instructions that make up the genetic inheritance. -

Guide to Interpreting Genomic Reports: a Genomics Toolkit
Guide to Interpreting Genomic Reports: A Genomics Toolkit A guide to genomic test results for non-genetics providers Created by the Practitioner Education Working Group of the Clinical Sequencing Exploratory Research (CSER) Consortium Genomic Report Toolkit Authors Kelly East, MS, CGC, Wendy Chung MD, PhD, Kate Foreman, MS, CGC, Mari Gilmore, MS, CGC, Michele Gornick, PhD, Lucia Hindorff, PhD, Tia Kauffman, MPH, Donna Messersmith , PhD, Cindy Prows, MSN, APRN, CNS, Elena Stoffel, MD, Joon-Ho Yu, MPh, PhD and Sharon Plon, MD, PhD About this resource This resource was created by a team of genomic testing experts. It is designed to help non-geneticist healthcare providers to understand genomic medicine and genome sequencing. The CSER Consortium1 is an NIH-funded group exploring genomic testing in clinical settings. Acknowledgements This work was conducted as part of the Clinical Sequencing Exploratory Research (CSER) Consortium, grants U01 HG006485, U01 HG006485, U01 HG006546, U01 HG006492, UM1 HG007301, UM1 HG007292, UM1 HG006508, U01 HG006487, U01 HG006507, R01 HG006618, and U01 HG007307. Special thanks to Alexandria Wyatt and Hugo O’Campo for graphic design and layout, Jill Pope for technical editing, and the entire CSER Practitioner Education Working Group for their time, energy, and support in developing this resource. Contents 1 Introduction and Overview ................................................................ 3 2 Diagnostic Results Related to Patient Symptoms: Pathogenic and Likely Pathogenic Variants . 8 3 Uncertain Results -
DNA Sequencing – How and Why Important
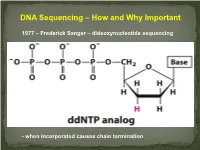
DNA Sequencing – How and Why Important 1977 – Frederick Sanger – dideoxynucleotide sequencing - when incorporated causes chain termination Original Sanger Sequencing Automated Sanger Sequencing Human Genome Project 1990 – funded as 15 year project to determine the nucleotide sequence of the entire human genome 2000 – Rough draft made available 2003 – The project was completed after final editing. Cost: $3 Billion - Identified approximately 23,000 protein coding genes - Fewer than 7 % of proteins are vertebrate specific - Complete data stored on multiple internet sites with tools for visualizing and searching UNDER currents BY KATHARINE MILLER IS CLINICAL GENOMICS TESTING WORTH IT? Cost-efectiveness studies yield answers to the complex question of whether clinical genomics testing has value. hole-genome testing has now variant and the possible benefts of test- to the right patient at the right time. reached the long-anticipated ing—are uncertain. Sequencing also Cost-efectiveness analyses are revealing W “$1,000 genome” level; and provides information about many difer- valuable benefts for certain patients, in more targeted genetic panels cost even ent genes, and each variant will have a the areas of rare pediatric disease, cancer, less. But the costs associated with diferent cost-beneft ratio. “You can’t do and pharmacogenomics. But the jury genomic testing don’t end with sequenc- a holistic view of the full beneft of these is still out as to whether whole exome ing. Additional expenditures—for follow- tests,” says Eman Biltaji, PhD, gradu- or whole genome sequencing (WES or up testing or treatments—may far exceed ate research assistant at the University of WGS) for healthy patients is worth it. -

DNA Polymerases Drive DNA Sequencing-By-Synthesis Technologies: Both Past and Present
View metadata, citation and similar papers at core.ac.uk brought to you by CORE REVIEW ARTICLEprovided by Frontiers - Publisher Connector published: 24 June 2014 doi: 10.3389/fmicb.2014.00305 DNA polymerases drive DNA sequencing-by-synthesis technologies: both past and present Cheng-Yao Chen* Protein Engineering Group, Illumina, San Diego, CA, USA Edited by: Next-generation sequencing (NGS) technologies have revolutionized modern biological and Andrew F.Gardner, New England biomedical research. The engines responsible for this innovation are DNA polymerases; Biolabs, USA they catalyze the biochemical reaction for deriving template sequence information. In fact, Reviewed by: DNA polymerase has been a cornerstone of DNA sequencing from the very beginning. Suleyman Yildirim, Walter Reed Army Institute of Research, USA Escherichia coli DNA polymerase I proteolytic (Klenow) fragment was originally utilized Andreas Marx, University of in Sanger’s dideoxy chain-terminating DNA sequencing chemistry. From these humble Konstanz, Germany beginnings followed an explosion of organism-specific, genome sequence information *Correspondence: accessible via public database. Family A/B DNA polymerases from mesophilic/thermophilic Cheng-Yao Chen, Protein Engineering bacteria/archaea were modified and tested in today’s standard capillary electrophoresis Group, Illumina, 5200 Illumina Way, San Diego, CA 92122, USA (CE) and NGS sequencing platforms.These enzymes were selected for their efficient incor- e-mail: [email protected] poration of bulky dye-terminator and reversible dye-terminator nucleotides respectively. Third generation, real-time single molecule sequencing platform requires slightly different enzyme properties. Enterobacterial phage φ29 DNA polymerase copies long stretches of DNA and possesses a unique capability to efficiently incorporate terminal phosphate- labeled nucleoside polyphosphates. -

Minds-On 4-Dna-Sequencing.Pdf
DNA BARCODING ACTION PROJECT Fish Market Survey BIOTECHNOLOGY MINDS-ON 4: DNA Sequencing Suggested Timing: 30 minutes In this hands-on activity, students will create a paper-based model of the process of DNA sequencing using the method established by Frederick Sanger in 1977 to understand how DNA is sequenced for procedures such as DNA barcoding. Prior Knowledge and Skills Success Criteria For each group of 4 students Understanding of basic mechanics of DNA replication Quality responses are BLM M5: DNA Sequencing (i.e., strand separation, annealing of primer, extension of provided during group 1 copy of DNA Sequencing – G (Guanine) DNA strand using DNA polymerase and termination) discussions 1 copy of DNA Sequencing – C (Cytosine) Understanding of complementary base pairing (C-G, A-T) Sequences correctly 1 copy of DNA Sequencing – T (Thymine) match base pair Familiarity with process of gel electrophoresis (migration 1 copy of DNA Sequencing – A (Adenine) template strand and of DNA fragments in a gel exposed to an electrical complementary 1 copy of DNA Sequencing – Group current) strand Consolidation For each group of 4 students Did you know? 4 pairs of scissors 1 green highlighter pen or marker Frederick Sanger won his second Nobel Prize for Chemistry in 1 stapler 1 roll transparent tape 1980, sharing it with Walter Gilbert, for their contributions 1 pink highlighter pen or marker concerning the determination of base sequences in nucleic 1 yellow highlighter pen or marker acids, and Paul Berg for his work on recombinant DNA. 1 blue highlighter pen or marker 1. Explain to the students that they will be determining the base sequence of a piece of DNA by modelling the process of DNA sequencing through the chain termination method developed by Fred Sanger in 1977.