Generations of Sequencing Technologies: from First to Next
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

DNA Sequencing and Sorting: Identifying Genetic Variations
BioMath DNA Sequencing and Sorting: Identifying Genetic Variations Student Edition Funded by the National Science Foundation, Proposal No. ESI-06-28091 This material was prepared with the support of the National Science Foundation. However, any opinions, findings, conclusions, and/or recommendations herein are those of the authors and do not necessarily reflect the views of the NSF. At the time of publishing, all included URLs were checked and active. We make every effort to make sure all links stay active, but we cannot make any guaranties that they will remain so. If you find a URL that is inactive, please inform us at [email protected]. DIMACS Published by COMAP, Inc. in conjunction with DIMACS, Rutgers University. ©2015 COMAP, Inc. Printed in the U.S.A. COMAP, Inc. 175 Middlesex Turnpike, Suite 3B Bedford, MA 01730 www.comap.com ISBN: 1 933223 71 5 Front Cover Photograph: EPA GULF BREEZE LABORATORY, PATHO-BIOLOGY LAB. LINDA SHARP ASSISTANT This work is in the public domain in the United States because it is a work prepared by an officer or employee of the United States Government as part of that person’s official duties. DNA Sequencing and Sorting: Identifying Genetic Variations Overview Each of the cells in your body contains a copy of your genetic inheritance, your DNA which has been passed down to you, one half from your biological mother and one half from your biological father. This DNA determines physical features, like eye color and hair color, and can determine susceptibility to medical conditions like hypertension, heart disease, diabetes, and cancer. -

The Bio Revolution: Innovations Transforming and Our Societies, Economies, Lives
The Bio Revolution: Innovations transforming economies, societies, and our lives economies, societies, our and transforming Innovations Revolution: Bio The Executive summary The Bio Revolution Innovations transforming economies, societies, and our lives May 2020 McKinsey Global Institute Since its founding in 1990, the McKinsey Global Institute (MGI) has sought to develop a deeper understanding of the evolving global economy. As the business and economics research arm of McKinsey & Company, MGI aims to help leaders in the commercial, public, and social sectors understand trends and forces shaping the global economy. MGI research combines the disciplines of economics and management, employing the analytical tools of economics with the insights of business leaders. Our “micro-to-macro” methodology examines microeconomic industry trends to better understand the broad macroeconomic forces affecting business strategy and public policy. MGI’s in-depth reports have covered more than 20 countries and 30 industries. Current research focuses on six themes: productivity and growth, natural resources, labor markets, the evolution of global financial markets, the economic impact of technology and innovation, and urbanization. Recent reports have assessed the digital economy, the impact of AI and automation on employment, physical climate risk, income inequal ity, the productivity puzzle, the economic benefits of tackling gender inequality, a new era of global competition, Chinese innovation, and digital and financial globalization. MGI is led by three McKinsey & Company senior partners: co-chairs James Manyika and Sven Smit, and director Jonathan Woetzel. Michael Chui, Susan Lund, Anu Madgavkar, Jan Mischke, Sree Ramaswamy, Jaana Remes, Jeongmin Seong, and Tilman Tacke are MGI partners, and Mekala Krishnan is an MGI senior fellow. -

Genomic Sequencing of Infectious Pathogens
Science, Technology Assessment, MARCH 2021 and Analytics WHY THIS MATTERS Genomic sequencing reveals the genetic code of an infectious disease pathogen, such as SARS-CoV-2, the SCIENCE & TECH SPOTLIGHT: virus that causes COVID-19. Newer, faster, and less costly sequencing can now be used to more quickly track GENOMIC SEQUENCING OF transmission, detect new variants, and develop vaccines and other countermeasures. However, challenges such INFECTIOUS PATHOGENS as high startup costs and privacy concerns remain. /// THE TECHNOLOGY How mature is it? First-generation sequencing is often used to confirm results of NGS. NGS is relatively new to the public health field, but is used What is it? Genomic sequencing technologies decode a pathogen’s to augment surveillance (i.e., data collection and analysis) for SARS- genetic material by identifying the order of chemical “letters” of its DNA CoV-2 in the U.S. and overseas. New technologies are allowing greater (or RNA, its chemical equivalent in some viruses). Each of four letters access to sequencing capabilities by making NGS portable, faster, and represents a chemical unit called a base. The sequence of the bases can more affordable (the cost of one sequence run is now one-millionth of reveal useful information for combatting disease. For example, sequencing what it was two decades ago). Genomic sequencing technologies enable of SARS-CoV-2 is used to track the spread of various strains, known as many different areas of infectious disease study. For example, they enable variants (see fig. 1). genomic epidemiology—the science of using pathogen genomic data to determine the distribution and spread of an infectious disease in a group How does it work? Technologies for sequencing genomes of pathogens of people or animals, and the application of this information to respond to use chemicals to break up the DNA or RNA into small fragments. -

DNA Sequencing with Thermus Aquaticus DNA Polymerase
Proc. Natl. Acad. Sci. USA Vol. 85, pp. 9436-9440, December 1988 Biochemistry DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA (thermophilic DNA polymerase/chain-termination/processivity/automation) MICHAEL A. INNIS*, KENNETH B. MYAMBO, DAVID H. GELFAND, AND MARY ANN D. BROW Department of Microbial Genetics, Cetus Corporation, 1400 Fifty-Third Street, Emeryville, CA 94608 Communicated by Hamilton 0. Smith, September 8, 1988 (receivedfor review August 15, 1988) ABSTRACT The highly thermostable DNA polymerase properties of Taq DNA polymerase that pertain to its advan- from Thermus aquaticus (Taq) is ideal for both manual and tages for DNA sequencing and its fidelity in PCR. automated DNA sequencing because it is fast, highly proces- sive, has little or no 3'-exonuclease activity, and is active over a broad range of temperatures. Sequencing protocols are MATERIALS presented that produce readable extension products >1000 Enzymes. Polynucleotide kinase from T4-infected Esche- bases having uniform band intensities. A combination of high richia coli cells was purchased from Pharmacia. Taq DNA reaction temperatures and the base analog 7-deaza-2'- polymerase, a single subunit enzyme with relative molecular deoxyguanosine was used to sequence through G+C-rich DNA mass of 94 kDa (specific activity, 200,000 units/mg; 1 unit and to resolve gel compressions. We modified the polymerase corresponds to 10 nmol of product synthesized in 30 min with chain reaction (PCR) conditions for direct DNA sequencing of activated salmon sperm DNA), was purified from Thermus asymmetric PCR products without intermediate purification aquaticus, strain YT-1 (ATCC no. -

Sanger Sequencing – a Hands-On Simulation Background And
Sanger sequencing – a hands-on simulation Background and Guidelines for Instructor Jared Young Correspondence concerning this article should be addressed to Jared Young, Mills College 5000 MacArthur Blvd, Oakland, CA 94613 Contact: [email protected] Synopsis This hands-on simulation teaches the Sanger (dideoxy) method of DNA sequencing. In the process of carrying out the exercise, students also confront DNA synthesis, especially as it relates to chemical structure, and the stochastic nature of biological processes. The exercise is designed for an introductory undergraduate genetics course for biology majors. The exercise can be completed in around 90-minutes, which can be broken up into a 50-minute period for the simulation and a follow-up 50-minute (or less) period for discussion. This follow-up could also take place in a Teaching Assistant (TA) led section. The exercise involves interactions between student pairs and the entire class. There is an accompanying student handout with prompts that should be invoked where indicated in these instructions. Introduction Sanger sequencing is an important technique: it revolutionized the field of Genetics and is still in wide use today. Sanger sequencing is a powerful pedagogical tool well-suited for inducing multiple “aha” moments: in achieving a deep understanding of the technique, students gain a better understanding of DNA and nucleotide structure, DNA synthesis, the stochastic nature of biological processes, the utility of visible chemical modifications (in this case, fluorescent dyes), gel electrophoresis, and the connection between a physical molecule and the information it contains. Sanger sequencing is beautiful: a truly elegant method that can bring a deep sense of satisfaction when it is fully understood. -

The Genomics Era: the Future of Genetics in Medicine - Glossary
The Genomics Era: the Future of Genetics in Medicine - Glossary The glossary below provides a list of key terms used throughout the course. You do not need to read them all now; we’ll be linking back to the main glossary step wherever these terms appear, so you may refer back to this list if you are unsure of the terminology being used. Term Definition The process of matching reads back to their original Alignment position in the reference genome. An allele is one of a number of alternative forms of the same gene or genetic locus. We inherit one copy Allele of our genetic code from our mother and one copy of our genetic code from our father. Each copy is known as an allele. Microarray based genomic comparative hybridisation. This is a technique used to detect chromosome imbalances by comparing patient and control DNA and comparing differences between the two sets. It is Array CGH a useful technique for detecting small chromosome deletions and duplications which would not have been detected with more traditional karyotyping techniques. A unit of DNA. There are four bases which form the Base cross links (or rungs) of the DNA double helix: adenine (A), thymine (T), guanine (G) and cytosine (C). Capture see Target enrichment. The process by which a cell becomes specialized in Cell differentiation order to perform a specific function. Centromere The point at which the sister chromatids are joined. #1 FutureLearn A structure located in the nucleus all living cells, comprised of DNA bound around proteins called histones. The normal number of chromosomes in each Chromosome human cell nucleus is 46 and is composed of 22 pairs of autosomes and a pair of sex chromosomes which determine gender: males have an X and a Y chromosome whilst females have two X chromosomes. -

DNA Sequencing) Time Binding Sites and Other Functional Sites Using Sequencing As a Means of Evaluating Chromatin Immunoprecipitation (Chip) Elaine R
COMMENTARY for re-sequencing genomes. With next- generation-based applications, we rapidly A brief history of will begin to annotate the human genome with the positions of regulatory protein- (DNA sequencing) time binding sites and other functional sites using sequencing as a means of evaluating chromatin immunoprecipitation (ChIP) Elaine R. Mardis experiments. A variant of ChIP will eluci- date how genome-wide patterns of histone Each spring, I co-teach a course to under- binding and DNA methylation relate to graduates that aims to introduce them to gene-expression regulation in various cell the brief history of genome sequencing states, such as differentiation or disease. and then immerses them in its current The short read-lengths of next-generation practices. Looking out at their eager faces, platforms also are ideal for discovering the I try (perhaps in vain) to capture for them sequences of non-coding RNA genes that in a 1 hour lecture what has been my life’s cannot be elucidated by in silico methods. focus for the past 20 years or so — DNA Bioinformatics-based analysis of genome- sequencing. If forced to summarize this wide DNA copy number, DNA-sequence time period succinctly, I would say, “Never variation and RNA-expression data sets, all a dull moment!” This is largely owing to the produced by next-generation sequencers, technological advances that have catapulted will be integrated to stitch together the genome sequencing from a cottage industry pieces of the biological ‘story’ being told to a high-throughput enterprise, akin to by genomic data. These stories will, in a factory. -

12.2% 122000 135M Top 1% 154 4800
View metadata, citation and similar papers at core.ac.uk brought to you by CORE We are IntechOpen, provided by IntechOpen the world’s leading publisher of Open Access books Built by scientists, for scientists 4,800 122,000 135M Open access books available International authors and editors Downloads Our authors are among the 154 TOP 1% 12.2% Countries delivered to most cited scientists Contributors from top 500 universities Selection of our books indexed in the Book Citation Index in Web of Science™ Core Collection (BKCI) Interested in publishing with us? Contact [email protected] Numbers displayed above are based on latest data collected. For more information visit www.intechopen.com 1 DNA Representation Bharti Rajendra Kumar B.T. Kumaon Institute of Technology, Dwarahat,Almora, Uttarakhand, India 1. Introduction The term DNA sequencing refers to methods for determining the order of the nucleotides bases adenine,guanine,cytosine and thymine in a molecule of DNA. The first DNA sequence were obtained by academic researchers,using laboratories methods based on 2- dimensional chromatography in the early 1970s. By the development of dye based sequencing method with automated analysis,DNA sequencing has become easier and faster. The knowledge of DNA sequences of genes and other parts of the genome of organisms has become indispensable for basic research studying biological processes, as well as in applied fields such as diagnostic or forensic research. DNA is the information store that ultimately dictates the structure of every gene product, delineates every part of the organisms. The order of the bases along DNA contains the complete set of instructions that make up the genetic inheritance. -

Cloning, DNA Amplification.Pdf
Molecular Biology DNA structure and function Recombinant DNA technology DNA amplification DNA sequencing Arthur Günzl, PhD Dept. of Genetics & Genome Sciences University of Connecticut Health Center Bioinformatics and Computational Biology Course (BME5800) September 13, 2016 Deoxyribonucleic Acid (DNA) 5’ 3’ 2’ The haploid human genome consists of ~3,000,000,000 bp The combined a-helical length is ~2 m A human nucleus’ diameter is ~10 mm, containing 46 chromosomes From Alberts et al., Molecular Biology of the Cell Which one is the reverse primer ? 5’-TTGGGAAGCTCCTTGTCA-3’ |||||||||||||||||| 3’-AACCCTTCGAGGAACAGT-5’ a. 5’-ACTGTTCCTCGAAGGGTT-3’ b. 5’-AACCCTTCGAGGAACAGT-3’ c. 5’-TGACAAGGAGCTTCCCAA-3’ d. 5’-GGTCCTGGAGAAAAGTCT-3’ e. None of the above Recombinant DNA Technology Recombination Process in which DNA molecules are broken and the fragments are rejoined in new combinations Recombinant DNA Any DNA molecule formed by joining DNA segments from different sources Recombinant DNA Technology Key Enzymes Restriction enzyme One of a large number of endonucleases that can cleave a DNA molecule at any site where a specific short nucleo- tide sequence occurs 5’-...GTACCTAGAATTCTT CTAG...-3’ ||||||||||||||||| 3’-...CATGGATCTTAAGTT GATC...-5’ Eco RI (sticky end) 5’-...GTACCTAG AATTCCTAGTG...-3’ |||||||| ||||||| 3’-...CATGGATCTTAA GGATCAC...-5’ SmaI CCC GGG Serratia marcescens Recombinant DNA Technology Key Enzymes DNA ligase Enzyme that joins the ends of two DNA strands together with a covalent bond to make a continuous DNA strand 5’-...GTACCTACCC-3’ 5’-pGGGCTAGTG...-3’ |||||||||| ||||||||| 3’-...CATGGATGGGp-5’ 3’-CCCGATCAC...-5’ T4 DNA ligase + ATP 5’-...GTACCTACCCGGGCTAGTG...-3’ ||||||||||||||||||| 3’-...CATGGATGGGCCCGATCAC...-5’ Recombinant DNA Technology plasmid cloning vector Multiple Cloning Site Gene cloning gene plasmid donor DNA vector RESTRICTION DIGEST DNA LIGASE (ATP) recombinant plasmid Transformation of E. -

Guide to Interpreting Genomic Reports: a Genomics Toolkit
Guide to Interpreting Genomic Reports: A Genomics Toolkit A guide to genomic test results for non-genetics providers Created by the Practitioner Education Working Group of the Clinical Sequencing Exploratory Research (CSER) Consortium Genomic Report Toolkit Authors Kelly East, MS, CGC, Wendy Chung MD, PhD, Kate Foreman, MS, CGC, Mari Gilmore, MS, CGC, Michele Gornick, PhD, Lucia Hindorff, PhD, Tia Kauffman, MPH, Donna Messersmith , PhD, Cindy Prows, MSN, APRN, CNS, Elena Stoffel, MD, Joon-Ho Yu, MPh, PhD and Sharon Plon, MD, PhD About this resource This resource was created by a team of genomic testing experts. It is designed to help non-geneticist healthcare providers to understand genomic medicine and genome sequencing. The CSER Consortium1 is an NIH-funded group exploring genomic testing in clinical settings. Acknowledgements This work was conducted as part of the Clinical Sequencing Exploratory Research (CSER) Consortium, grants U01 HG006485, U01 HG006485, U01 HG006546, U01 HG006492, UM1 HG007301, UM1 HG007292, UM1 HG006508, U01 HG006487, U01 HG006507, R01 HG006618, and U01 HG007307. Special thanks to Alexandria Wyatt and Hugo O’Campo for graphic design and layout, Jill Pope for technical editing, and the entire CSER Practitioner Education Working Group for their time, energy, and support in developing this resource. Contents 1 Introduction and Overview ................................................................ 3 2 Diagnostic Results Related to Patient Symptoms: Pathogenic and Likely Pathogenic Variants . 8 3 Uncertain Results -
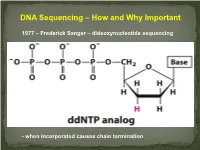
DNA Sequencing – How and Why Important
DNA Sequencing – How and Why Important 1977 – Frederick Sanger – dideoxynucleotide sequencing - when incorporated causes chain termination Original Sanger Sequencing Automated Sanger Sequencing Human Genome Project 1990 – funded as 15 year project to determine the nucleotide sequence of the entire human genome 2000 – Rough draft made available 2003 – The project was completed after final editing. Cost: $3 Billion - Identified approximately 23,000 protein coding genes - Fewer than 7 % of proteins are vertebrate specific - Complete data stored on multiple internet sites with tools for visualizing and searching UNDER currents BY KATHARINE MILLER IS CLINICAL GENOMICS TESTING WORTH IT? Cost-efectiveness studies yield answers to the complex question of whether clinical genomics testing has value. hole-genome testing has now variant and the possible benefts of test- to the right patient at the right time. reached the long-anticipated ing—are uncertain. Sequencing also Cost-efectiveness analyses are revealing W “$1,000 genome” level; and provides information about many difer- valuable benefts for certain patients, in more targeted genetic panels cost even ent genes, and each variant will have a the areas of rare pediatric disease, cancer, less. But the costs associated with diferent cost-beneft ratio. “You can’t do and pharmacogenomics. But the jury genomic testing don’t end with sequenc- a holistic view of the full beneft of these is still out as to whether whole exome ing. Additional expenditures—for follow- tests,” says Eman Biltaji, PhD, gradu- or whole genome sequencing (WES or up testing or treatments—may far exceed ate research assistant at the University of WGS) for healthy patients is worth it. -

Glossary of Terms
GLOSSARY OF TERMS Table of Contents A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z A Amino acids: any of a class of 20 molecules that are combined to form proteins in living things. The sequence of amino acids in a protein and hence protein function are determined by the genetic code. From http://www.geneticalliance.org.uk/glossary.htm#C • The building blocks of proteins, there are 20 different amino acids. From https://www.yourgenome.org/glossary/amino-acid Antisense: Antisense nucleotides are strings of RNA or DNA that are complementary to "sense" strands of nucleotides. They bind to and inactivate these sense strands. They have been used in research, and may become useful for therapy of certain diseases (See Gene silencing). From http://www.encyclopedia.com/topic/Antisense_DNA.aspx. Antisense and RNA interference are referred as gene knockdown technologies: the transcription of the gene is unaffected; however, gene expression, i.e. protein synthesis (translation), is lost because messenger RNA molecules become unstable or inaccessible. Furthermore, RNA interference is based on naturally occurring phenomenon known as Post-Transcriptional Gene Silencing. From http://www.ncbi.nlm.nih.gov/probe/docs/applsilencing/ B Biobank: A biobank is a large, organised collection of samples, usually human, used for research. Biobanks catalogue and store samples using genetic, clinical, and other characteristics such as age, gender, blood type, and ethnicity. Some samples are also categorised according to environmental factors, such as whether the donor had been exposed to some substance that can affect health.