Title Generation and Maintenance of Species Diversity in Leaf Cone Moths
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Experience 2,000 Follow the Path from the Roundabout Towards East, Past the Small Lake and You Will Be at the Main Entrance
The Arboretum in Hørsholm Why an Arboretum? Te Arboretum is a living collection of more than 8,500 trees Less than a hundred species of trees, shrubs and woody climbers and shrubs. Te objective of the collection is to include and are native to Denmark. Te main reason for this is the fact that observe woody plant species that can survive and grow under most of Northern Europe during each of the last 7 major glacia- Danish conditions. tions was covered by ice, and the unglaciated landscapes north of the Alps were covered by arctic and sub-arctic tundras and steppes. All plants in the Arboretum have detailed documentation of age, Te rich fora in e.g. North America and Eastern Asia could escape GPS location, species identity, and the geographic origin of where such cold and dry climates. seeds were collected. Te information is stored in a database and can be accessed from the Arboretum webpage. In spite of our poor native dendrofora, a rich variety of exotic species and provenances are able to grow in the Danish climate. Te collection is managed by the Department of Geosciences At present the collections of the Arboretum include approximately Cornus kousa and Natural Resource Management, University of Copenhagen. 2,000 species, subspecies and cultivars in an area of 25 ha. Te Arboretum is a continuation of the Forest Botanical Garden in reveals if the seed was collected from a natural population (W), Charlottenlund from 1838. Te establishment of the Arboretum in from a plant that originates from a natural population (Z) Hrsholm was initiated in 1936, to support teaching and research or is a cultivated variety (G). -

Arboretum News Armstrong News & Featured Publications
Georgia Southern University Digital Commons@Georgia Southern Arboretum News Armstrong News & Featured Publications Spring 2019 Arboretum News Georgia Southern University- Armstrong Campus Follow this and additional works at: https://digitalcommons.georgiasouthern.edu/armstrong-arbor-news Part of the Education Commons This article is brought to you for free and open access by the Armstrong News & Featured Publications at Digital Commons@Georgia Southern. It has been accepted for inclusion in Arboretum News by an authorized administrator of Digital Commons@Georgia Southern. For more information, please contact [email protected]. Arboretum News Issue 9 | Spring 2019 A Newsletter of the Georgia Southern University Armstrong Campus Arboretum From the Editor: Arboretum News, published by the Grounds Operations Department ’d like to introduce you to the Armstrong Arboretum of the new of Georgia Southern University- IGeorgia Southern University-Armstrong Campus. Designated Armstrong Campus, is distributed as an on-campus arboretum in 2001 by former Armstrong to faculty, staff, students and Atlantic State University president Dr. Thomas Jones, the friends of the Armstrong Arboretum. The Arboretum university recognized the rich diversity of plant life on campus. encompasses Armstrong’s 268- The Arboretum continues to add to that diversity and strives to acre campus and displays a wide function as a repository for the preservation and the conservation variety of shrubs and other woody of plants from all over the world. We also hope to inspire students, plants. Developed areas of campus faculty, staff and visitors to appreciate the incredible diversity contain native and introduced species of trees and shrubs. Most that plants have to offer. -

Acer Miyabei
Woody Plants Database [http://woodyplants.cals.cornell.edu] Species: Acer miyabei (ay'ser mi-YA-bee-eye) Miyabe Maple Cultivar Information * See specific cultivar notes on next page. Ornamental Characteristics Size: Tree < 30 feet Height: 35'-45', Width: 30' Leaves: Deciduous Shape: upright oval to rounded, can have open or dense branching, low branching Ornamental Other: prefers full sun, tolerates partial shade Environmental Characteristics Light: Full sun, Part shade Hardy To Zone: 5a Soil Ph: Can tolerate acid to alkaline soil (pH 5.0 to 8.0) CU Structural Soil™: Yes Insect Disease none of significance Bare Root Transplanting Easy Other easy to transplant B&B or < 2.5" caliper bare root. Native to Japan Moisture Tolerance 1 Woody Plants Database [http://woodyplants.cals.cornell.edu] Occasionally saturated Consistently moist, Occasional periods of Prolonged periods of or very wet soil well-drained soil dry soil dry soil 1 2 3 4 5 6 7 8 9 10 11 12 2 Woody Plants Database [http://woodyplants.cals.cornell.edu] Cultivars for Acer miyabei Showing 1-2 of 2 items. Cultivar Name Notes Rugged Ridge 'Rugged Ridge' - more deeply furrowed corky bark than species State Street 'State Street' (a.k.a. Morton) - hardy to zone 4; upright oval form; good uniform branching; dark green foliage; good golden yellow fall color; possibly fast growing 3 Woody Plants Database [http://woodyplants.cals.cornell.edu] Photos Acer miyabei trunk Acer miyabei foliage 4 Woody Plants Database [http://woodyplants.cals.cornell.edu] Acer miyabei habit Acer miyabei - Bark 5 Woody Plants Database [http://woodyplants.cals.cornell.edu] Acer miyabei - Leaf Acer miyabei - Habit 6. -
Environmental Factors Controlling the Distribution of Forest Plants with Special Reference to Floral Mixture in the Boreo-Nemora
Environmental Factors Controlling the Distribution of Forest Plants with Special Reference to Floral Mixture in the Title Boreo-Nemoral Ecotone, Hokkaido Island Author(s) Uemura, Shigeru Environmental science, Hokkaido University : journal of the Graduate School of Environmental Science, Hokkaido Citation University, Sapporo, 15(2), 1-54 Issue Date 1993-03-25 Doc URL http://hdl.handle.net/2115/37276 Type bulletin (article) File Information 15(2)_1-54.pdf Instructions for use Hokkaido University Collection of Scholarly and Academic Papers : HUSCAP 1 Environ,Sci.,I'Iokl{aidoUniversity 15(2) 1-54 Dec,1992 Environmental Factors Controlling the Distribution of Forest PlaRts with Special Reference to Floral Mixture in the Boreo-Nemoral EcotoRe, Hokkaido Island Shigeru Uemura Department of Biosystem Management, Division of Environmental Conservation, Graduate School of Environmental Science, Hokkaiclo University, Sapporo 060, Japan Abstract Effects of climatic factors on the plant distribution were examined by means of direct gradient analysis, and the relationship of forest flora with Iife form and phytogeographical distribution was exaniined. Subsequently, leaf phenology of forest plants were analyzed to evaluate the adaptive signifi- cance in relation to the environments in forest understory. In the boreo-nemoral forest ecotone, Kokkaido Island, northern Japan, co-occurrence of northern and southern plants in a certain forest site is more notable in the understory than in the crown, and this dates back to the late--Quaternary period, where the decrease in temperature associated with the glacial period forced the unclerstory plants to adapt their life forms or leaf habits to snowcover and to light conditions of the interior forests, I<ey words: Direct gradient analysis; Floral mixture; Leaf phenology; Mixed forest; Phytogoegraphy; Snowcover; Understory Intoroduetion In the upper-middle latitudes of Europe, eastern Asia and eastern North America, the boreal coniferous forest formation confronts to the temperate hardwood forest formation. -

Acer Buergerianum Plants, Adequately Moist in Summer but Well Drained in Winter Is "Trident Maple" a Pretty Small Tree Whose Grace Is Enhanced by the Key to Success
Acer buergerianum plants, adequately moist in summer but well drained in winter is "Trident Maple" A pretty small tree whose grace is enhanced by the key to success. 3m. the small three-lobed leaves. Particularly good autumn colour begins scarlet turning orange-yellow. A good hardy Maple Acer x conspicuum 'Silver Cardinal' tolerant of many less favoured sites. 4m. This Snakebark has the most incredible pink and cream variegated foliage, highlighted by the red petioles and young stems. It Acer circinatum 'Monroe' occurred as a chance seedling of A. pensylvanicum and received A plant I've lusted after for years! Shrubby habit, with deeply an Award of Merit in 1985. Our stock is directly derived from incised light green leaves (even more so than A. japonicum the original seedling in the Windsor Great Park. Unless your soil 'Aconitifolium'). Predominantly yellow autumn colours may is very good, it is safest in dappled shade. 3m. develop some orange. Worthy of a special site. 3m. Acer x conspicuum 'Silver Vein' Acer circinatum 'Pacific Fire' A hybrid between A. davidii George Forrest and A. Imagine the coral bark colour of Acer palmatum 'Sangokaku' pensylvanicum Erythrocladum found at Hilliers about 1960. It is combined with the larger leaves and more tolerant growth arguably the best of the basic snakebarks for garden suitability requirements of this species, and the result is a plant with and good colour with its rich purple and white striped winter awesome potential. Fantastic autumn colour too. bark, becoming green with maturity. 5m. Acer circinatum 'Sunglow' Acer davidii This has been on my "wanted" list ever since I first saw it Delightful small tree noted for dazzling autumn colour and photographed! Apricot coloured young growth matures to attractive white striped purple bark in winter. -

Maples in the Landscape Sheriden Hansen, Jaydee Gunnell, and Andra Emmertson
EXTENSION.USU.EDU Maples in the Landscape Sheriden Hansen, JayDee Gunnell, and Andra Emmertson Introduction Maple trees (Acer sp.) are a common fixture and beautiful addition to Utah landscapes. There are over one hundred species, each with numerous cultivars (cultivated varieties) that are native to both North America and much of Northern Europe. Trees vary in size and shape, from small, almost prostrate forms like certain Japanese maples (Acer palmatum) and shrubby bigtooth maples (Acer grandidentatum) to large and stately shade trees like the Norway maple (Acer platanoides). Tree shape can vary greatly, ranging from upright, columnar, rounded, pyramidal to spreading. Because trees come in a Figure 1. Severe iron chlorosis on maple. Note the range of shapes and sizes, there is almost always a interveinal chlorosis characterized by the yellow leaves spot in a landscape that can be enhanced by the and green veins. Spotting on the leaves is indicative of the addition of a maple. Maples can create a focal point beginning of tissue necrosis from a chronic lack of iron. and ornamental interest in the landscape, providing interesting textures and colors, and of course, shade. some micronutrients, particularly iron, to be less Fall colors typically range from yellow to bright red, available, making it difficult for certain trees to take adding a burst of color to the landscape late in the up needed nutrients. A common problem associated season. with maples in the Intermountain West is iron chlorosis (Figure 1). This nutrient deficiency causes Recommended Cultivars yellowing leaves (chlorosis) with green veins, and in extreme conditions, can cause death of leaf edges. -
Botanical Name Common Name
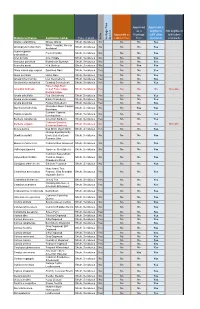
Approved Approved & as a eligible to Not eligible to Approved as Frontage fulfill other fulfill other Type of plant a Street Tree Tree standards standards Heritage Tree Tree Heritage Species Botanical Name Common name Native Abelia x grandiflora Glossy Abelia Shrub, Deciduous No No No Yes White Forsytha; Korean Abeliophyllum distichum Shrub, Deciduous No No No Yes Abelialeaf Acanthropanax Fiveleaf Aralia Shrub, Deciduous No No No Yes sieboldianus Acer ginnala Amur Maple Shrub, Deciduous No No No Yes Aesculus parviflora Bottlebrush Buckeye Shrub, Deciduous No No No Yes Aesculus pavia Red Buckeye Shrub, Deciduous No No Yes Yes Alnus incana ssp. rugosa Speckled Alder Shrub, Deciduous Yes No No Yes Alnus serrulata Hazel Alder Shrub, Deciduous Yes No No Yes Amelanchier humilis Low Serviceberry Shrub, Deciduous Yes No No Yes Amelanchier stolonifera Running Serviceberry Shrub, Deciduous Yes No No Yes False Indigo Bush; Amorpha fruticosa Desert False Indigo; Shrub, Deciduous Yes No No No Not eligible Bastard Indigo Aronia arbutifolia Red Chokeberry Shrub, Deciduous Yes No No Yes Aronia melanocarpa Black Chokeberry Shrub, Deciduous Yes No No Yes Aronia prunifolia Purple Chokeberry Shrub, Deciduous Yes No No Yes Groundsel-Bush; Eastern Baccharis halimifolia Shrub, Deciduous No No Yes Yes Baccharis Summer Cypress; Bassia scoparia Shrub, Deciduous No No No Yes Burning-Bush Berberis canadensis American Barberry Shrub, Deciduous Yes No No Yes Common Barberry; Berberis vulgaris Shrub, Deciduous No No No No Not eligible European Barberry Betula pumila -

Latitudinal Gradient in Leaf Defense Traits of Woody Plants Along Japanese Archipelago
Latitudinal gradient in leaf defense traits of woody plants along Japanese archipelago 日本産樹木種における、葉防御形質の緯度傾度 Saihanna 1 General Introduction It is estimated that over the twenty million species of organisms are living on our planet, and all of these organisms adapted to their own living environment, namely niche (Hatchinson 1957). Not only the abiotic factors but biotic interaction plays a key role in the maintenance of biodiversity. Animal-plant interactions are one of the most important topic in community ecology (e.g. Morin 1999). Plants and herbivore insects have accounted for about half of the entire diversity on the earth (Strong et al., 1984). Plant-herbivore interactions are extremely complex, which should lead the tremendous diversity of both plants and herbivores (e.g. Gutierrez et al., 1984; Hay et al., 1989). Although the interaction between these two components, namely co-speciation, should account for this diversification, most of the studies so far, tend to explain this interaction only from one side of them. Plants have interacted with insect herbivores for several hundred million years, which should lead to complex defense systems against various herbivores (Fürstenberg-Hägg et al., 2013). This interaction between plants and herbivores has long proposed the opportunity for studying the mechanism of the creation and maintenance of biological diversity because of its universality and generality (Strong et al. 1984; Ali and Agrawal 2012). It is believed that the evolution of plant defense traits followed by counter-adaptations in herbivores could lead to bursts of adaptive radiation of both components (Ehrlich and Raven 1969). Understanding the coevolution of plant and insect species and macroevolution of adaptive traits has inspired biologists for some decades, yet has been challenging to study even present days (Schluter, 2000). -

Number 3, Spring 1998 Director’S Letter
Planning and planting for a better world Friends of the JC Raulston Arboretum Newsletter Number 3, Spring 1998 Director’s Letter Spring greetings from the JC Raulston Arboretum! This garden- ing season is in full swing, and the Arboretum is the place to be. Emergence is the word! Flowers and foliage are emerging every- where. We had a magnificent late winter and early spring. The Cornus mas ‘Spring Glow’ located in the paradise garden was exquisite this year. The bright yellow flowers are bright and persistent, and the Students from a Wake Tech Community College Photography Class find exfoliating bark and attractive habit plenty to photograph on a February day in the Arboretum. make it a winner. It’s no wonder that JC was so excited about this done soon. Make sure you check of themselves than is expected to seedling selection from the field out many of the special gardens in keep things moving forward. I, for nursery. We are looking to propa- the Arboretum. Our volunteer one, am thankful for each and every gate numerous plants this spring in curators are busy planting and one of them. hopes of getting it into the trade. preparing those gardens for The magnolias were looking another season. Many thanks to all Lastly, when you visit the garden I fantastic until we had three days in our volunteers who work so very would challenge you to find the a row of temperatures in the low hard in the garden. It shows! Euscaphis japonicus. We had a twenties. There was plenty of Another reminder — from April to beautiful seven-foot specimen tree damage to open flowers, but the October, on Sunday’s at 2:00 p.m. -

Morton Arboretum Bulletin of Popular Information
VOL. 13 No. 4 APR IL, 1938 MORTON ARBORETUM JOY MORTON · FOUNDER BULLETIN OF LI SLE, ILLINOIS POPULAR INFORMATION JOURNAL OF A TOUR OF ARBORETA, BOTANICAL GARDENS AND NURSERIES OF NORTHERN EUROPE PART 2 ARBORETUM OF TI-IE STATE AGRICULTURAL COLLEGE, Wageningen, Netherlands Acreage : 15. Notes: collections rather ex tensive, well cared for. Aeswlus Pavia rosea nana (Dwarf Red Buckeye) Crataegus monogyna pendula (Weeping English Hawthorn) Prwws Petunnikowi (Turkestan Almond) Prwws serotina pendula (Weeping Black Cherry) Pterocarya fraxinifolia dwnosa (Shrubby Caucasian Walnut) Querws robur laurifolia (Laurel-leaved Engli sh Oak) Tilia platyphyllos nana (Dwarf Bigleaf European Linden) BOTANIC GARDEN OF TI-IE STATE UNIVERSITY OF UTRECHT, Utrecht (Baarn), Netherlands Established: 1920. Acreage: 10. Celtis jessoensis (Hackberry of Japanese and Korean origin) Crataegomespilus Gillotii (hybrid of Crataegus monogyna and Mespilus germanica) Liriodendron Tulipifera variegata (Variegated Tulip Tree) Suaeda fruticosa (low, heath.like shrub of saline habitats) BOTANIC GARDEN OF TI-IE UN IVERSITY OF AMSTERDAM, Amsterdam, Netherlands Established: 1682. Acreage: 4. N ates: collection rather small; large speci· mens of Corylus colurna (Turkish Hazel) , Davidia Vilmoriniana (Dove Tree) and Rhus vernici/liw (Lacquer Tree) . Aralia chinensis alba marginata (White margined Chin ese Aralia) Aralia chinensis aiireo marginata (Yellow margined Chinese Aralia) Crataegus monogyna /l exuosa (English Hawthorn with curiously twisted branches) Deutzia discolor maculosa (Chinese Deutzia variety ) BOTANICAL GARDEN OF TI-IE STATE UN IV ERSITY, Leiden, Netherlands Established:! 1587. This garden ranks as the fifth oldest in Europe. It originally covered only one·sixth of an acre and was laid out in 1594 by the noted botanist, Clusiu s (Charles l'Escluse) . -

October 1961 , Volume 40, Number 4 305
TIIE .A:M:ERICA.N ~GAZINE AMERICAN HORTICULTURAL SOCIETY A union of the Amej'ican HOTticu~tural Society and the AmeTican HOTticultural Council 1600 BLADENSBURG ROAD, NORTHEAST. WASHINGTON 2, D. C. For United Horticulture *** to accumulate, increase, and disseminate horticultural intOTmation B. Y. MORRISON, Editor Directors Terms Expiring 1961 JAMES R. HARLOW, Managing Editor STUART M. ARMSTRONG Maryland Editorial Committee JOH N L. CREECH . Maryland W. H . HODGE, Chairman WILLIAM H. FREDERICK, JR. Delaware JOH N L. CREECH FRANCIS PATTESON-KNIGHT FREDERIC P. LEE Virginia DONALD WYMAN CONRAD B. LINK Massachusetts CURTIS MAY T erms Expiring 1962 FREDERICK G . MEYER FREDERIC P. LEE WILBUR H . YOUNGMAN Maryland HENRY T . SKINNER District of Columbia OfJiceTS GEORGE H. SPALDING California PRESIDENT RICHARD P. WHITE DONAlJD WYMAN Distj'ict of Columbia Jamaica Plain, Massachusetts ANNE WERTSNER WOOD Pennsylvania FIRST VICE· PRESIDENT Ternu Expiring 1963 ALBERT J . IRVING New l'm'k, New York GRETCHEN HARSHBARGER Iowa SECOND VICE-PRESIDENT MARY W. M. HAKES Maryland ANNE WERTSNER W ' OOD FREDERIC HEUTTE Swarthmore, Pennsylvania Virginia W . H. HODGE SECRETARY-TREASURER OLIVE E. WEATHERELL ALBERT J . IRVING Washington, D, C. New York The Ame"ican Horticultural Magazine is the official publication of the American Horticultural Society and is issued four times a year during the quarters commencing with January, April, July and October. It is devoted to the dissemination of knowledge in the science and art of growing ornamental plants, fruits, vegetables, and related subjects. Original papers increasing the historical, varietal, and cultural knowledges of plant materials of economic and aesthetic importance are welcomed and will be published as early as possible. -

Download PCN-Acer-2017-Holdings.Pdf
PLANT COLLECTIONS NETWORK MULTI-INSTITUTIONAL ACER LIST 02/13/18 Institutional NameAccession no.Provenance* Quan Collection Id Loc.** Vouchered Plant Source Acer acuminatum Wall. ex D. Don MORRIS Acer acuminatum 1994-009 W 2 H&M 1822 1 No Quarryhill BG, Glen Ellen, CA QUARRYHILL Acer acuminatum 1993.039 W 4 H&M1822 1 Yes Acer acuminatum 1993.039 W 1 H&M1822 1 Yes Acer acuminatum 1993.039 W 1 H&M1822 1 Yes Acer acuminatum 1993.039 W 1 H&M1822 1 Yes Acer acuminatum 1993.076 W 2 H&M1858 1 No Acer acuminatum 1993.076 W 1 H&M1858 1 No Acer acuminatum 1993.139 W 1 H&M1921 1 No Acer acuminatum 1993.139 W 1 H&M1921 1 No UBCBG Acer acuminatum 1994-0490 W 1 HM.1858 0 Unk Sichuan Exp., Kew BG, Howick Arb., Quarry Hill ... Acer acuminatum 1994-0490 W 1 HM.1858 0 Unk Sichuan Exp., Kew BG, Howick Arb., Quarry Hill ... Acer acuminatum 1994-0490 W 1 HM.1858 0 Unk Sichuan Exp., Kew BG, Howick Arb., Quarry Hill ... UWBG Acer acuminatum 180-59 G 1 1 Yes National BG, Glasnevin Total of taxon 18 Acer albopurpurascens Hayata IUCN Red List Status: DD ATLANTA Acer albopurpurascens 20164176 G 1 2 No Crug Farm Nursery QUARRYHILL Acer albopurpurascens 2003.088 U 1 1 No Total of taxon 2 Acer amplum (Gee selection) DAWES Acer amplum (Gee selection) D2014-0117 G 1 1 No Gee Farms, Stockbridge, MI 49285 Total of taxon 1 Acer amplum 'Gold Coin' DAWES Acer amplum 'Gold Coin' D2015-0013 G 1 2 No Gee Farms, Stockbridge, MI 49285, USA Acer amplum 'Gold Coin' D2017-0075 G 2 2 No Shinn, Edward T., Wall Township, NJ 07719-9128 Total of taxon 3 Acer argutum Maxim.