Bat-Associated Diseases Now Available
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Recognition TLR7 Signaling Beyond Endosomal Dendritic Cells
Flavivirus Activation of Plasmacytoid Dendritic Cells Delineates Key Elements of TLR7 Signaling beyond Endosomal Recognition This information is current as of September 29, 2021. Jennifer P. Wang, Ping Liu, Eicke Latz, Douglas T. Golenbock, Robert W. Finberg and Daniel H. Libraty J Immunol 2006; 177:7114-7121; ; doi: 10.4049/jimmunol.177.10.7114 http://www.jimmunol.org/content/177/10/7114 Downloaded from References This article cites 38 articles, 21 of which you can access for free at: http://www.jimmunol.org/content/177/10/7114.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on September 29, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2006 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Flavivirus Activation of Plasmacytoid Dendritic Cells Delineates Key Elements of TLR7 Signaling beyond Endosomal Recognition1 Jennifer P. Wang,2* Ping Liu,† Eicke Latz,* Douglas T. Golenbock,* Robert W. Finberg,* and Daniel H. -

Living with Wildlife - Flying Foxes Fact Sheet No
LIVING WITH WILDLIFE - FLYING FOXES FACT SHEET NO. 0063 Living with wildlife - Flying Foxes What are flying foxes? flying foxes use to mark their territory and to attract females during the mating season. Flying foxes, also known as fruit bats, are winged mammals belonging to the sub-order group of megabats. Unlike the smaller insectivorous microbats, the Do flying foxes carry diseases? animal navigates using their eye sight and smell, as opposed to echolocation, Like most wildlife and pets, flying foxes may carry diseases that can affect and feed on nectar, pollen and fruit. Flying foxes forage from over 100 humans. Australian Bat Lyssavirus can be transmitted directly from flying species of native plants and may supplement this diet with introduced plants foxes to humans. The risk of contracting Lyssavirus is extremely low, with found in gardens, orchards and urban areas. transmission only possible through direct contact of saliva from an infected Of the four species of flying foxes native to mainland Australia, three reside animal with a skin penetrating bite or scratch. in the Gladstone Region. These species include the grey-headed flying fox Flying foxes are natural hosts of the Hendra virus, however, there is no (Pteropus poliocephalus), black flying fox (Pteropus alecto) and little red evidence that the virus can be transmitted directly to humans. It is believed flying fox (Pteropus scapulatus). that the virus is transmitted from flying foxes to horses through exposure to All of these species are protected under the Nature Conservation Act urine or birthing fluids. Vaccination is the most effective way of reducing the 1992 and the grey-headed flying fox is also listed as ‘vulnerable’ under the risk of the virus infecting horses. -

A Preliminary Study of Viral Metagenomics of French Bat Species in Contact with Humans: Identification of New Mammalian Viruses
A preliminary study of viral metagenomics of French bat species in contact with humans: identification of new mammalian viruses. Laurent Dacheux, Minerva Cervantes-Gonzalez, Ghislaine Guigon, Jean-Michel Thiberge, Mathias Vandenbogaert, Corinne Maufrais, Valérie Caro, Hervé Bourhy To cite this version: Laurent Dacheux, Minerva Cervantes-Gonzalez, Ghislaine Guigon, Jean-Michel Thiberge, Mathias Vandenbogaert, et al.. A preliminary study of viral metagenomics of French bat species in contact with humans: identification of new mammalian viruses.. PLoS ONE, Public Library of Science, 2014, 9 (1), pp.e87194. 10.1371/journal.pone.0087194.s006. pasteur-01430485 HAL Id: pasteur-01430485 https://hal-pasteur.archives-ouvertes.fr/pasteur-01430485 Submitted on 9 Jan 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License A Preliminary Study of Viral Metagenomics of French Bat Species in Contact with Humans: Identification of New Mammalian Viruses Laurent Dacheux1*, Minerva Cervantes-Gonzalez1, -

Daytime Behaviour of the Grey-Headed Flying Fox Pteropus Poliocephalus Temminck (Pteropodidae: Megachiroptera) at an Autumn/Winter Roost
DAYTIME BEHAVIOUR OF THE GREY-HEADED FLYING FOX PTEROPUS POLIOCEPHALUS TEMMINCK (PTEROPODIDAE: MEGACHIROPTERA) AT AN AUTUMN/WINTER ROOST K.A. CONNELL, U. MUNRO AND F.R. TORPY Connell KA, Munro U and Torpy FR, 2006. Daytime behaviour of the grey-headed flying fox Pteropus poliocephalus Temminck (Pteropodidae: Megachiroptera) at an autumn/winter roost. Australian Mammalogy 28: 7-14. The grey-headed flying fox (Pteropus poliocephalus Temminck) is a threatened large fruit bat endemic to Australia. It roosts in large colonies in rainforest patches, mangroves, open forest, riparian woodland and, as native habitat is reduced, increasingly in vegetation within urban environments. The general biology, ecology and behaviour of this bat remain largely unknown, which makes it difficult to effectively monitor, protect and manage this species. The current study provides baseline information on the daytime behaviour of P. poliocephalus in an autumn/winter roost in urban Sydney, Australia, between April and August 2003. The most common daytime behaviours expressed by the flying foxes were sleeping (most common), grooming, mating/courtship, and wing spreading (least common). Behaviours differed significantly between times of day and seasons (autumn and winter). Active behaviours (i.e., grooming, mating/courtship, wing spreading) occurred mainly in the morning, while sleeping predominated in the afternoon. Mating/courtship and wing spreading were significantly higher in April (reproductive period) than in winter (non-reproductive period). Grooming was the only behaviour that showed no significant variation between sample periods. These results provide important baseline data for future comparative studies on the behaviours of flying foxes from urban and ‘natural’ camps, and the development of management strategies for this species. -

Pdf Available
Virology 554 (2021) 89–96 Contents lists available at ScienceDirect Virology journal homepage: www.elsevier.com/locate/virology Diverse cressdnaviruses and an anellovirus identifiedin the fecal samples of yellow-bellied marmots Anthony Khalifeh a, Daniel T. Blumstein b,**, Rafaela S. Fontenele a, Kara Schmidlin a, C´ecile Richet a, Simona Kraberger a, Arvind Varsani a,c,* a The Biodesign Center for Fundamental and Applied Microbiomics, School of Life Sciences, Center for Evolution and Medicine, Arizona State University, Tempe, AZ, 85287, USA b Department of Ecology & Evolutionary Biology, Institute of the Environment & Sustainability, University of California Los Angeles, Los Angeles, CA, 90095, USA c Structural Biology Research Unit, Department of Clinical Laboratory Sciences, University of Cape Town, 7925, Cape Town, South Africa ARTICLE INFO ABSTRACT Keywords: Over that last decade, coupling multiple strand displacement approaches with high throughput sequencing have Marmota flaviventer resulted in the identification of genomes of diverse groups of small circular DNA viruses. Using a similar Anelloviridae approach but with recovery of complete genomes by PCR, we identified a diverse group of single-stranded vi Genomoviridae ruses in yellow-bellied marmot (Marmota flaviventer) fecal samples. From 13 fecal samples we identified viruses Cressdnaviricota in the family Genomoviridae (n = 7) and Anelloviridae (n = 1), and several others that ware part of the larger Cressdnaviricota phylum but not within established families (n = 19). There were also circular DNA molecules identified (n = 4) that appear to encode one viral-like gene and have genomes of <1545 nts. This study gives a snapshot of viruses associated with marmots based on fecal sampling. -

Survival of Human Norovirus Surrogates in Juices and Their Inactivation Using Novel Methods
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Masters Theses Graduate School 5-2011 Survival of Human Norovirus Surrogates In Juices and their Inactivation Using Novel Methods Katie Marie Horm [email protected] Follow this and additional works at: https://trace.tennessee.edu/utk_gradthes Recommended Citation Horm, Katie Marie, "Survival of Human Norovirus Surrogates In Juices and their Inactivation Using Novel Methods. " Master's Thesis, University of Tennessee, 2011. https://trace.tennessee.edu/utk_gradthes/882 This Thesis is brought to you for free and open access by the Graduate School at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Masters Theses by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. To the Graduate Council: I am submitting herewith a thesis written by Katie Marie Horm entitled "Survival of Human Norovirus Surrogates In Juices and their Inactivation Using Novel Methods." I have examined the final electronic copy of this thesis for form and content and recommend that it be accepted in partial fulfillment of the equirr ements for the degree of Master of Science, with a major in Food Science and Technology. Doris H. D'Souza, Major Professor We have read this thesis and recommend its acceptance: Federico M. Harte, Gina M. Pighetti Accepted for the Council: Carolyn R. Hodges Vice Provost and Dean of the Graduate School (Original signatures are on file with official studentecor r ds.) Survival of Human Norovirus Surrogates In Juices and their Inactivation Using Novel Methods A Thesis Presented for the Master of Science Degree The University of Tennessee, Knoxville Katie Marie Horm May 2011 Acknowledgments I would like to think my major professor/advisor Dr. -

2020 Taxonomic Update for Phylum Negarnaviricota (Riboviria: Orthornavirae), Including the Large Orders Bunyavirales and Mononegavirales
Archives of Virology https://doi.org/10.1007/s00705-020-04731-2 VIROLOGY DIVISION NEWS 2020 taxonomic update for phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales Jens H. Kuhn1 · Scott Adkins2 · Daniela Alioto3 · Sergey V. Alkhovsky4 · Gaya K. Amarasinghe5 · Simon J. Anthony6,7 · Tatjana Avšič‑Županc8 · María A. Ayllón9,10 · Justin Bahl11 · Anne Balkema‑Buschmann12 · Matthew J. Ballinger13 · Tomáš Bartonička14 · Christopher Basler15 · Sina Bavari16 · Martin Beer17 · Dennis A. Bente18 · Éric Bergeron19 · Brian H. Bird20 · Carol Blair21 · Kim R. Blasdell22 · Steven B. Bradfute23 · Rachel Breyta24 · Thomas Briese25 · Paul A. Brown26 · Ursula J. Buchholz27 · Michael J. Buchmeier28 · Alexander Bukreyev18,29 · Felicity Burt30 · Nihal Buzkan31 · Charles H. Calisher32 · Mengji Cao33,34 · Inmaculada Casas35 · John Chamberlain36 · Kartik Chandran37 · Rémi N. Charrel38 · Biao Chen39 · Michela Chiumenti40 · Il‑Ryong Choi41 · J. Christopher S. Clegg42 · Ian Crozier43 · John V. da Graça44 · Elena Dal Bó45 · Alberto M. R. Dávila46 · Juan Carlos de la Torre47 · Xavier de Lamballerie38 · Rik L. de Swart48 · Patrick L. Di Bello49 · Nicholas Di Paola50 · Francesco Di Serio40 · Ralf G. Dietzgen51 · Michele Digiaro52 · Valerian V. Dolja53 · Olga Dolnik54 · Michael A. Drebot55 · Jan Felix Drexler56 · Ralf Dürrwald57 · Lucie Dufkova58 · William G. Dundon59 · W. Paul Duprex60 · John M. Dye50 · Andrew J. Easton61 · Hideki Ebihara62 · Toufc Elbeaino63 · Koray Ergünay64 · Jorlan Fernandes195 · Anthony R. Fooks65 · Pierre B. H. Formenty66 · Leonie F. Forth17 · Ron A. M. Fouchier48 · Juliana Freitas‑Astúa67 · Selma Gago‑Zachert68,69 · George Fú Gāo70 · María Laura García71 · Adolfo García‑Sastre72 · Aura R. Garrison50 · Aiah Gbakima73 · Tracey Goldstein74 · Jean‑Paul J. Gonzalez75,76 · Anthony Grifths77 · Martin H. Groschup12 · Stephan Günther78 · Alexandro Guterres195 · Roy A. -
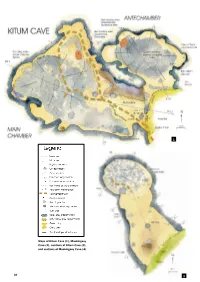
30 SWARA October – December 2007 Maps of Kitum Cave
1 Maps of Kitum Cave (1), Mackingeny Cave (2), sections of Kitum Cave (3), and sections of Mackingeny Cave (4). 30 SWARA October – December 2007 2 behaviour 3 Mount Elgon’s ‘elephant caves’ Joyce Lundberg and Donald McFarlane contemplate and map the extraordinary underground attractions of western Kenya’s Mount Elgon National Park. 4 SWARA October – December 2007 31 ne of Kenya’s least vis- that are far too busy eating the herbivore ited parks, the Mount equivalent of chocolate mousse to take Elgon National Park much notice of their potential fate. Oprotects a narrow band The salts are mainly mirabalite, of forest climbing the eastern flank sodium sulphate (commonly called of East Africa’s fifth highest mas- Glauber’s salt), which grows out sif, standing 4,321 metres (14,178 from the walls and can form curved crys- feet) above sea level (see map below). tals resembling pig’s tusks. The crystals are For most visitors, the Park’s principal licked off the wall by buffalos or scraped/ attraction rests in its suite of unique caves, gouged out by bushbuck and elephants. of which the Kitum and the Makingeny Buffalos cannot scrape the rock themselves, Caves are best known. Used by the Elgony so they eat mainly the leftover bits dropped people for centuries, the Elgon caves came by the elephants. The marks left by bush- to wider attention through the writings of buck teeth and elephant tusks are clearly Joseph Thomson (Through Masai Land, identifiable on the cave walls. 1885), and are thought to have been the These marks should not be confused inspiration too for H. -

First Description of Adenovirus, Enterovirus, Rotavirus and Torque
First description of Adenovirus, Enterovirus, Rotavirus and Torque teno virus in water samples collected from the Arroio Dilúvio, Porto Alegre, Brazil Vecchia, AD.a,b, Fleck, JD.a,b, Comerlato, J.c, Kluge, M.b, Bergamaschi, B.c, Da Silva, JVS.b, Da Luz, RB.b, Teixeira, TF.b, Garbinatto, GN.d, Oliveira, DV.d, Zanin, JG.d, Van der Sand, S.d, Frazzon, APG.d, Franco, AC.c, Roehe, PM.c,e and Spilki, FR.a,b* aPrograma de Pós-Graduação em Qualidade Ambiental, Universidade Feevale, CEP 93352-000, Novo Hamburgo, RS, Brazil bLaboratório de Microbiologia Molecular, Instituto de Ciências da Saúde, Universidade Feevale, CEP 93352-000, Novo Hamburgo, RS, Brazil cLaboratório de Virologia, Departamento de Microbiologia, Instituto de Ciências Básicas da Saúde, Universidade Federal do Rio Grande do Sul – UFRGS, Av. Sarmento Leite, 500, CEP 90050-170, Porto Alegre, RS, Brazil dDepartamento de Microbiologia, Instituto de Ciências Básicas da Saúde, Universidade Federal do Rio Grande do Sul – UFRGS, Av. Sarmento Leite, 500, CEP 90050-170, Porto Alegre, RS, Brazil eInstituto de Pesquisa Veterinária “Desidério Finamor” – IPVDF, Fundação Estadual de Pesquisa Agropecuária – FEPAGRO-Saúde Animal, Estrada do Conde, 6000, CEP 92990-000, Eldorado do Sul, RS, Brazil *e-mail: [email protected] Received May 11, 2011 – Accepted July 14, 2011 – Distributed May 31, 2012 (With 1 figure) Abstract Adenovirus (AdV), enterovirus (EV), genogroup A rotaviruses (GARV) and Torque teno virus (TTV) are non-enveloped viral agents excreted in feces and so may contaminate water bodies. In the present study, the molecular detection of these viruses was performed in samples of surface water collected from the Arroio Dilúvio, a waterstream that crosses the city of Porto Alegre, RS, Brazil, receiving great volumes of non-treated sewage from a large urban area. -

Characterizing and Evaluating the Zoonotic Potential of Novel Viruses Discovered in Vampire Bats
viruses Article Characterizing and Evaluating the Zoonotic Potential of Novel Viruses Discovered in Vampire Bats Laura M. Bergner 1,2,* , Nardus Mollentze 1,2 , Richard J. Orton 2 , Carlos Tello 3,4, Alice Broos 2, Roman Biek 1 and Daniel G. Streicker 1,2 1 Institute of Biodiversity, Animal Health and Comparative Medicine, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow G12 8QQ, UK; [email protected] (N.M.); [email protected] (R.B.); [email protected] (D.G.S.) 2 MRC–University of Glasgow Centre for Virus Research, Glasgow G61 1QH, UK; [email protected] (R.J.O.); [email protected] (A.B.) 3 Association for the Conservation and Development of Natural Resources, Lima 15037, Peru; [email protected] 4 Yunkawasi, Lima 15049, Peru * Correspondence: [email protected] Abstract: The contemporary surge in metagenomic sequencing has transformed knowledge of viral diversity in wildlife. However, evaluating which newly discovered viruses pose sufficient risk of infecting humans to merit detailed laboratory characterization and surveillance remains largely speculative. Machine learning algorithms have been developed to address this imbalance by ranking the relative likelihood of human infection based on viral genome sequences, but are not yet routinely Citation: Bergner, L.M.; Mollentze, applied to viruses at the time of their discovery. Here, we characterized viral genomes detected N.; Orton, R.J.; Tello, C.; Broos, A.; through metagenomic sequencing of feces and saliva from common vampire bats (Desmodus rotundus) Biek, R.; Streicker, D.G. and used these data as a case study in evaluating zoonotic potential using molecular sequencing Characterizing and Evaluating the data. -

Taxonomy of the Order Bunyavirales: Update 2019
Archives of Virology (2019) 164:1949–1965 https://doi.org/10.1007/s00705-019-04253-6 VIROLOGY DIVISION NEWS Taxonomy of the order Bunyavirales: update 2019 Abulikemu Abudurexiti1 · Scott Adkins2 · Daniela Alioto3 · Sergey V. Alkhovsky4 · Tatjana Avšič‑Županc5 · Matthew J. Ballinger6 · Dennis A. Bente7 · Martin Beer8 · Éric Bergeron9 · Carol D. Blair10 · Thomas Briese11 · Michael J. Buchmeier12 · Felicity J. Burt13 · Charles H. Calisher10 · Chénchén Cháng14 · Rémi N. Charrel15 · Il Ryong Choi16 · J. Christopher S. Clegg17 · Juan Carlos de la Torre18 · Xavier de Lamballerie15 · Fēi Dèng19 · Francesco Di Serio20 · Michele Digiaro21 · Michael A. Drebot22 · Xiaˇoméi Duàn14 · Hideki Ebihara23 · Toufc Elbeaino21 · Koray Ergünay24 · Charles F. Fulhorst7 · Aura R. Garrison25 · George Fú Gāo26 · Jean‑Paul J. Gonzalez27 · Martin H. Groschup28 · Stephan Günther29 · Anne‑Lise Haenni30 · Roy A. Hall31 · Jussi Hepojoki32,33 · Roger Hewson34 · Zhìhóng Hú19 · Holly R. Hughes35 · Miranda Gilda Jonson36 · Sandra Junglen37,38 · Boris Klempa39 · Jonas Klingström40 · Chūn Kòu14 · Lies Laenen41,42 · Amy J. Lambert35 · Stanley A. Langevin43 · Dan Liu44 · Igor S. Lukashevich45 · Tāo Luò1 · Chuánwèi Lüˇ 19 · Piet Maes41 · William Marciel de Souza46 · Marco Marklewitz37,38 · Giovanni P. Martelli47 · Keita Matsuno48,49 · Nicole Mielke‑Ehret50 · Maria Minutolo3 · Ali Mirazimi51 · Abulimiti Moming14 · Hans‑Peter Mühlbach50 · Rayapati Naidu52 · Beatriz Navarro20 · Márcio Roberto Teixeira Nunes53 · Gustavo Palacios25 · Anna Papa54 · Alex Pauvolid‑Corrêa55 · Janusz T. Pawęska56,57 · Jié Qiáo19 · Sheli R. Radoshitzky25 · Renato O. Resende58 · Víctor Romanowski59 · Amadou Alpha Sall60 · Maria S. Salvato61 · Takahide Sasaya62 · Shū Shěn19 · Xiǎohóng Shí63 · Yukio Shirako64 · Peter Simmonds65 · Manuela Sironi66 · Jin‑Won Song67 · Jessica R. Spengler9 · Mark D. Stenglein68 · Zhèngyuán Sū19 · Sùróng Sūn14 · Shuāng Táng19 · Massimo Turina69 · Bó Wáng19 · Chéng Wáng1 · Huálín Wáng19 · Jūn Wáng19 · Tàiyún Wèi70 · Anna E. -

Emergence and Returning Activity in the Indian Flying Fox, Pteropus Giganteus (Chiroptera: Pteropodidae)
International Journal of Geography and Geology 1(1):1-9 International Journal of Geography and Geology journal homepage: http://aessweb.com/journal-detail.php?id=5011 EMERGENCE AND RETURNING ACTIVITY IN THE INDIAN FLYING FOX, PTEROPUS GIGANTEUS (CHIROPTERA: PTEROPODIDAE) M. R. Sudhakaran1 D. P. Swamidoss2 P. Parvathiraj3 ABSTRACT Approach: After a diurnal resting in the roost, bats adapts some behavioural pattern to get themselves active towards foraging there by involving in various activities. Their activity pattern differs from time to time depending on the change in climatic factors. But the behavioural activities they involved varies from time to time. Observation was done on the emergence and returning gate i. e the emergence or returning of first to last bat, pre emergence behaviour and post return behaviour, and influence of moonlight on foraging activity. Key Words: Post return activity, emergence gate, behaviour, pre-emergence behaviour. INTRODUCTION Most of the megachiropteran’s found in the sub tropics roost in tents, in manmade structures like old houses and temples and in foliages or hollows in trees, mainly to evade from the climatic factors, which affect them. Indian flying fox, Pteropus giganteus roosts in open foliages of trees, a peculiar character of this genus. Papers dealing with behavioural aspects of P. giganteus are scarce. Apart from Neuweiler (1969), other works on behavioural aspects focused on copulatory behaviour (Koilraj et al., 2001), roost preference (Acharya, 1936), mating (Bhatt, 1942), local migration (Breadow, 1931; Nelson, 1965a) and general ecology and biology (Brosset, 1962). Bats roosting in closed environments involved in various behavioural activities during their emergence (Kunz, 1982) like light sampling, flying inside its roost etc.