The Evolution of Cell Types in Animals: Emerging Principles from Molecular Studies
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

FGF Signaling in Gastrulation and Neural Development in Nematostella Vectensis, an Anthozoan Cnidarian
Dev Genes Evol DOI 10.1007/s00427-006-0122-3 ORIGINAL ARTICLE FGF signaling in gastrulation and neural development in Nematostella vectensis, an anthozoan cnidarian David Q. Matus & Gerald H. Thomsen & Mark Q. Martindale Received: 8 June 2006 /Accepted: 3 November 2006 # Springer-Verlag 2007 Abstract The fibroblast growth factor (FGF) signal trans- planula stages known as the apical tuft. These results duction pathway serves as one of the key regulators of early suggest a conserved role for FGF signaling molecules in metazoan development, displaying conserved roles in the coordinating both gastrulation and neural induction that specification of endodermal, mesodermal, and neural fates predates the Cambrian explosion and the origins of the during vertebrate development. FGF signals also regulate Bilateria. gastrulation, in part, by triggering epithelial to mesenchy- mal transitions in embryos of both vertebrates and Keywords Gastrulation . Neurogenesis . invertebrates. Thus, FGF signals coordinate gastrulation Evolution of development movements across many different phyla. To help under- stand the breadth of FGF signaling deployment across the animal kingdom, we have examined the presence and Introduction expression of genes encoding FGF pathway components in the anthozoan cnidarian Nematostella vectensis. We isolat- Fibroblast growth factors (FGFs) were originally isolated ed three FGF ligands (NvFGF8A, NvFGF8B,and from vertebrate brain and pituitary fibroblasts for their roles NvFGF1A), two FGF receptors (NvFGFRa and NvFGFRb), in angiogenesis, mitogenesis, cellular differentiation, mi- and two orthologs of vertebrate FGF responsive genes, gration, and tissue-injury repair (Itoh and Ornitz 2004; Sprouty (NvSprouty), an inhibitor of FGF signaling, and Ornitz and Itoh 2001; Popovici et al. 2005). FGFs signal Churchill (NvChurchill), a Zn finger transcription factor. -

Assembly of the Cnidarian Camera-Type Eye from Vertebrate-Like Components
Assembly of the cnidarian camera-type eye from vertebrate-like components Zbynek Kozmik*†, Jana Ruzickova*‡, Kristyna Jonasova*‡, Yoshifumi Matsumoto§, Pavel Vopalensky*, Iryna Kozmikova*, Hynek Strnad*, Shoji Kawamura§, Joram Piatigorsky†¶, Vaclav Paces*, and Cestmir Vlcek*† *Institute of Molecular Genetics, Academy of Sciences of the Czech Republic, Videnska 1083, 142 20 Prague 4, Czech Republic; §Graduate School of Frontier Sciences, University of Tokyo, 5-1-5 Kashiwanoha, Kashiwa, Chiba 277-8562, Japan; and ¶National Eye Institute, National Institutes of Health, Bethesda, MD 20892-3655 Edited by Eviatar Nevo, University of Haifa, Haifa, Israel, and approved April 11, 2008 (received for review January 14, 2008) Animal eyes are morphologically diverse. Their assembly, however, containing Tripedalia eyes have sophisticated visual optics as do always relies on the same basic principle, i.e., photoreceptors advanced bilaterian phyla (11). located in the vicinity of dark shielding pigment. Cnidaria as the In the present work, we characterize genes required for the likely sister group to the Bilateria are the earliest branching phylum assembly of camera-type eyes in Tripedalia. We show that the with a well developed visual system. Here, we show that camera- genetic building blocks typical of vertebrate eyes, namely ciliary type eyes of the cubozoan jellyfish, Tripedalia cystophora, use opsin and the melanogenic pathway, are used by the cubozoan genetic building blocks typical of vertebrate eyes, namely, a ciliary eyes. Although our findings of unsuspected parallelism are phototransduction cascade and melanogenic pathway. Our find- consistent with either an independent origin or common ances- ings indicative of parallelism provide an insight into eye evolution. try of cubozoan and vertebrate eyes, we believe the present data Combined, the available data favor the possibility that vertebrate favor the former alternative. -

Introduction to the Cell Cell History Cell Structures and Functions
Introduction to the cell cell history cell structures and functions CK-12 Foundation December 16, 2009 CK-12 Foundation is a non-profit organization with a mission to reduce the cost of textbook materials for the K-12 market both in the U.S. and worldwide. Using an open-content, web-based collaborative model termed the “FlexBook,” CK-12 intends to pioneer the generation and distribution of high quality educational content that will serve both as core text as well as provide an adaptive environment for learning. Copyright ©2009 CK-12 Foundation This work is licensed under the Creative Commons Attribution-Share Alike 3.0 United States License. To view a copy of this license, visit http://creativecommons.org/licenses/by-sa/3.0/us/ or send a letter to Creative Commons, 171 Second Street, Suite 300, San Francisco, California, 94105, USA. Contents 1 Cell structure and function dec 16 5 1.1 Lesson 3.1: Introduction to Cells .................................. 5 3 www.ck12.org www.ck12.org 4 Chapter 1 Cell structure and function dec 16 1.1 Lesson 3.1: Introduction to Cells Lesson Objectives • Identify the scientists that first observed cells. • Outline the importance of microscopes in the discovery of cells. • Summarize what the cell theory proposes. • Identify the limitations on cell size. • Identify the four parts common to all cells. • Compare prokaryotic and eukaryotic cells. Introduction Knowing the make up of cells and how cells work is necessary to all of the biological sciences. Learning about the similarities and differences between cell types is particularly important to the fields of cell biology and molecular biology. -

1 Eric Davidson and Deep Time Douglas H. Erwin Department Of
Eric Davidson and Deep Time Douglas H. Erwin Department of Paleobiology, MRC-121 National Museum of Natural History Washington, DC 20013-7012 E-mail: [email protected] Abstract Eric Davidson had a deep and abiding interest in the role developmental mechanisms played in the generating evolutionary patterns documented in deep time, from the origin of the euechinoids to the processes responsible for the morphological architectures of major animal clades. Although not an evolutionary biologist, Davidson’s interests long preceded the current excitement over comparative evolutionary developmental biology. Here I discuss three aspects at the intersection between his research and evolutionary patterns in deep time: First, understanding the mechanisms of body plan formation, particularly those associated with the early diversification of major metazoan clades. Second, a critique of early claims about ancestral metazoans based on the discoveries of highly conserved genes across bilaterian animals. Third, Davidson’s own involvement in paleontology through a collaborative study of the fossil embryos from the Ediacaran Doushantuo Formation in south China. Keywords Eric Davidson – Evolution – Gene regulatory networks – Bodyplan – Cambrian Radiation – Echinoderms 1 Introduction Eric Davidson was a developmental biologist, not an evolutionary biologist or paleobiologist. He was driven to understand the mechanisms of gene regulatory control and how they controlled development, but this focus was deeply embedded within concerns about the relationship between development and evolution. Questions about the origin of major metazoan architectures or body plans were central to Eric’s concerns since at least the late 1960s. His 1971 paper with Roy Britten includes a section on “The Evolutionary Growth of the Genome” illustrated with a figure depicting variations in genome size in major animal groups and a metazoan phylogeny (Britten and Davidson 1971). -
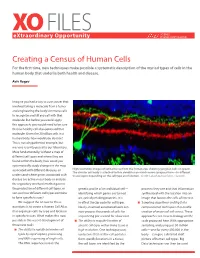
Creating a Census of Human Cells
XO FILES SCIENCE eXtraordinary Opportunity PHILANTHROPY ALLIANCE Creating a Census of Human Cells For the first time, new techniques make possible a systematic description of the myriad types of cells in the human body that underlie both health and disease. Aviv Regev Imagine you had a way to cure cancer that involved taking a molecule from a tumor and engineering the body’s immune cells to recognize and kill any cell with that molecule. But before you could apply this approach, you would need to be sure that no healthy cell also expressed that molecule. Given the 20 trillion cells in a human body, how would you do that? This is not a hypothetical example, but was one recently posed to our laboratory. More fundamentally, without a map of different cell types and where they are found within the body, how could you systematically study changes in the map associated with different diseases, or High resolution images of retinal tissue from the human eye, showing ganglion cells (in green). The circular cell body is attached to thin dendrites on which nerve synapses form—in different understand where genes associated with tissue layers depending on the cell type and function. Credit: Lab of Joshua Sanes, Harvard. disease are active in our body or analyze the regulatory mechanisms that govern the production of different cell types, or genetic profile of an individual cell— proteins they use and that information sort out how different cell types combine identifying which genes are turned synthesized with the location into an to form specific tissues? on, actively making proteins. -

Cadherin Switch Marks Germ Layer Formation in the Diploblastic Sea Anemone Nematostella Vectensis
bioRxiv preprint doi: https://doi.org/10.1101/488270; this version posted December 6, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Cadherin switch marks germ layer formation in the diploblastic sea anemone Nematostella vectensis PUKHLYAKOVA, E.A.1, KIRILLOVA, A.1,2, KRAUS, Y.A. 2, TECHNAU, U.1 1 Department for Molecular Evolution and Development, Centre of Organismal Systems Biology, University of Vienna, Althanstraße 14, A-1090 Vienna, Austria. 2 Department of Evolutionary Biology, Biological Faculty, Moscow State University, Leninskie Gory 1/12, 119991, Moscow, Russia. Key words: cadherin, cell adhesion, morphogenesis, germ layers, Nematostella, Cnidaria Abstract Morphogenesis is a shape-building process during development of multicellular organisms. During this process the establishment and modulation of cell-cell contacts play an important role. Cadherins, the major cell adhesion molecules, form adherens junctions connecting ephithelial cells. Numerous studies in Bilateria have shown that cadherins are associated with the regulation of cell differentiation, cell shape changes, cell migration and tissue morphogenesis. To date, the role of Cadherins in non- bilaterians is unknown. Here, we study the expression and the function of two paralogous classical cadherins, cadherin1 and cadherin3, in the diploblastic animal, the sea anemone Nematostella vectensis. We show that a cadherin switch is accompanying the formation of germ layers. Using specific antibodies, we show that both cadherins are localized to adherens junctions at apical and basal positions in ectoderm and endoderm. -

Robustness and Timing of Cellular Differentiation Through Population-Based Symmetry Breaking
AR TI CLE Robustness AND TIMING OF CELLULAR DIFFERENTIATION THROUGH population-based SYMMETRY BREAKING Angel StanoeV1 | Christian Schröter1 | Aneta KOSESKA1∗ 1Department OF Systemic Cell Biology, Max Planck Institute OF Molecular PhYSIOLOGY, Although COORDINATED BEHAVIOR OF MULTICELLULAR ORGANISMS Dortmund, GermanY RELIES ON INTERCELLULAR communication, THE DYNAMICAL mech- Correspondence ANISM THAT UNDERLIES SYMMETRY BREAKING IN HOMOGENEOUS Email: populations, THEREBY GIVING RISE TO HETEROGENEOUS CELL TYPES ∗[email protected] REMAINS UNCLEAR. In CONTRAST TO THE PREVALENT cell-intrinsic VIEW OF DIFFERENTIATION WHERE MULTISTABILITY ON THE SINGLE CELL LEVEL IS A NECESSARY pre-requisite, WE PROPOSE THAT het- EROGENEOUS CELLULAR IDENTITIES EMERGE AND ARE MAINTAINED COOPERATIVELY ON A POPULATION LEVel. Identifying A GENERIC SYMMETRY BREAKING mechanism, WE DEMONSTRATE THAT ROBUST PROPORTIONS BETWEEN DIFFERENTIATED CELL TYPES ARE INTRINSIC FEATURES OF THIS INHOMOGENEOUS STEADY STATE solution. Crit- ICAL ORGANIZATION IN THE VICINITY OF THE SYMMETRY BREAKING BIFURCATION ON THE OTHER hand, SUGGESTS THAT TIMING OF cel- LULAR DIFFERENTIATION EMERGES FOR GROWING POPULATIONS IN A self-organized MANNER. Setting A CLEAR DISTINCTION BETWEEN pre-eXISTING ASYMMETRIES AND SYMMETRY BREAKING EVents, WE THUS DEMONSTRATE THAT EMERGENT SYMMETRY breaking, IN CONJUNCTION WITH ORGANIZATION NEAR CRITICALITY NECESSARILY LEADS TO ROBUSTNESS AND ACCURACY DURING DEVelopment. KEYWORDS SYMMETRY breaking; differentiation; INHOMOGENEOUS STEADY STATE Abbreviations: -

Xenacoelomorpha's Significance for Understanding Bilaterian Evolution
Available online at www.sciencedirect.com ScienceDirect Xenacoelomorpha’s significance for understanding bilaterian evolution Andreas Hejnol and Kevin Pang The Xenacoelomorpha, with its phylogenetic position as sister biology models are the fruitfly Drosophila melanogaster and group of the Nephrozoa (Protostomia + Deuterostomia), plays the nematode Caenorhabditis elegans, in which basic prin- a key-role in understanding the evolution of bilaterian cell types ciples of developmental processes have been studied in and organ systems. Current studies of the morphological and great detail. It might be because the field of evolutionary developmental diversity of this group allow us to trace the developmental biology — EvoDevo — has its origin in evolution of different organ systems within the group and to developmental biology and not evolutionary biology that reconstruct characters of the most recent common ancestor of species under investigation are often called ‘model spe- Xenacoelomorpha. The disparity of the clade shows that there cies’. Criteria for selected representative species are cannot be a single xenacoelomorph ‘model’ species and primarily the ease of access to collected material and strategic sampling is essential for understanding the evolution their ability to be cultivated in the lab [1]. In some cases, of major traits. With this strategy, fundamental insights into the a supposedly larger number of ancestral characters or a evolution of molecular mechanisms and their role in shaping dominant role in ecosystems have played an additional animal organ systems can be expected in the near future. role in selecting model species. These arguments were Address used to attract sufficient funding for genome sequencing Sars International Centre for Marine Molecular Biology, University of and developmental studies that are cost-intensive inves- Bergen, Thormøhlensgate 55, 5008 Bergen, Norway tigations. -

A Concise Review on the Classification and Nomenclature of Stem Cells Kök Hücrelerinin S›N›Fland›R›Lmas› Ve Isimlendirilmesine Iliflkin K›Sa Bir Derleme
Review 57 A concise review on the classification and nomenclature of stem cells Kök hücrelerinin s›n›fland›r›lmas› ve isimlendirilmesine iliflkin k›sa bir derleme Alp Can Ankara University Medical School, Department of Histology and Embryology, Ankara, Turkey Abstract Stem cell biology and regenerative medicine is a relatively young field. However, in recent years there has been a tremen- dous interest in stem cells possibly due to their therapeutic potential in disease states. As a classical definition, a stem cell is an undifferentiated cell that can produce daughter cells that can either remain a stem cell in a process called self-renew- al, or commit to a specific cell type via the initiation of a differentiation pathway leading to the production of mature progeny cells. Despite this acknowledged definition, the classification of stem cells has been a perplexing notion that may often raise misconception even among stem cell biologists. Therefore, the aim of this brief review is to give a conceptual approach to classifying the stem cells beginning from the early morula stage totipotent embryonic stem cells to the unipotent tissue-resident adult stem cells, also called tissue-specific stem cells. (Turk J Hematol 2008; 25: 57-9) Key words: Stem cells, embryonic stem cells, tissue-specific stem cells, classification, progeny. Özet Kök hücresi biyolojisi ve onar›msal t›p görece yeni alanlard›r. Buna karfl›n, son y›llarda çeflitli hastal›klarda tedavi amac›yla kullan›labilme potansiyelleri nedeniyle kök hücrelerine ola¤anüstü bir ilgi art›fl› vard›r. Klasik tan›m›yla kök hücresi, kendini yenileme ad› verilen mekanizmayla farkl›laflmadan kendini ço¤altan veya bir dizi farkl›laflma aflamas›ndan geçerek olgun hücrelere dönüflebilen hücrelerdir. -

Bodily Complexity: Integrated Multicellular Organizations for Contraction-Based Motility
fphys-10-01268 October 11, 2019 Time: 16:13 # 1 HYPOTHESIS AND THEORY published: 15 October 2019 doi: 10.3389/fphys.2019.01268 Bodily Complexity: Integrated Multicellular Organizations for Contraction-Based Motility Argyris Arnellos1,2* and Fred Keijzer3 1 IAS-Research Centre for Life, Mind & Society, Department of Logic and Philosophy of Science, University of the Basque Country, San Sebastián, Spain, 2 Department of Product and Systems Design Engineering, Complex Systems and Service Design Lab, University of the Aegean, Syros, Greece, 3 Department of Theoretical Philosophy, University of Groningen, Groningen, Netherlands Compared to other forms of multicellularity, the animal case is unique. Animals—barring some exceptions—consist of collections of cells that are connected and integrated to such an extent that these collectives act as unitary, large free-moving entities capable of sensing macroscopic properties and events. This animal configuration Edited by: Matteo Mossio, is so well-known that it is often taken as a natural one that ‘must’ have evolved, UMR8590 Institut d’Histoire et given environmental conditions that make large free-moving units ‘obviously’ adaptive. de Philosophie des Sciences et des Techniques (IHPST), France Here we question the seemingly evolutionary inevitableness of animals and introduce Reviewed by: a thesis of bodily complexity: The multicellular organization characteristic for typical Stuart A. Newman, animals requires the integration of a multitude of intrinsic bodily features between its New York Medical -

Molecular Homology & the Ancient Genetic Toolkit: How Evolutionary
Regis University ePublications at Regis University All Regis University Theses Spring 2019 Molecular Homology & the Ancient Genetic Toolkit: How Evolutionary Development Could Shape Your Next Doctor's Appointment Elizabeth G. Plender Follow this and additional works at: https://epublications.regis.edu/theses Part of the Cell Biology Commons, Developmental Biology Commons, Evolution Commons, Molecular Biology Commons, and the Molecular Genetics Commons Recommended Citation Plender, Elizabeth G., "Molecular Homology & the Ancient Genetic Toolkit: How Evolutionary Development Could Shape Your Next Doctor's Appointment" (2019). All Regis University Theses. 905. https://epublications.regis.edu/theses/905 This Thesis - Open Access is brought to you for free and open access by ePublications at Regis University. It has been accepted for inclusion in All Regis University Theses by an authorized administrator of ePublications at Regis University. For more information, please contact [email protected]. MOLECULAR HOMOLOGY AND THE ANCIENT GENETIC TOOLKIT: HOW EVOLUTIONARY DEVELOPMENT COULD SHAPE YOUR NEXT DOCTOR’S APPOINTMENT A thesis submitted to Regis College The Honors Program In partial fulfillment of the requirements For Graduation with Honors By Elizabeth G. Plender May 2019 Thesis written by Elizabeth Plender Approved by Thesis Advisor Thesis Reader Accepted by Director, University Honors Program ii iii TABLE OF CONTENTS List of Figures ............................................................................................................................................... -

The Palaeoscolecida and the Evolution of the Ecdysozoa Andrey Yu
The Palaeoscolecida and the evolution of the Ecdysozoa Andrey Yu. Zhuravlev1, José Antonio Gámez Vintaned2 and Eladio Liñán1 1Área y Museo de Paleontología, Departamento de Ciencias de la Tierra, Facultad de Ciencias, Universidad de Zaragoza, C/ Pedro Cerbuna, 12, E-50009 Zaragoza, Spain 2Área de Paleontología, Departamento de Geologica, Facultad de Ciencias Biológicas, Univeristat de València, C/ Doctor Moliner, 50, E-46100 Burjassot, Spain Email: [email protected] AbstrAct rÉsUMÉ Palaeoscolecidans are a key group for understanding the ear- Les Paléoscolécides sont un groupe clé pour la compréhen- ly evolution of the Ecdysozoa. The Palaeoscolecida possess sion des débuts de l’évolution des Ecdysozoa. Les Pal- a terminal mouth and an anus, an invertible proboscis with aeoscolecida possèdent une bouche terminale et un anus, pointed scalids, a thick integument of diverse plates, sensory un proboscis inversible aux scalides pointues, un tégument papillae and caudal hooks. These are features that draw a épais de plaques diverses, des papilles sensorielles et des secret out of these worms, indicating palaeoscolecidan af- crochets caudaux. Ceux-ci sont des traits qui tirent un secret finities with the phylum Cephalorhyncha, which embraces de ces vers, ce qui indique des affinités paléoscolecides avec priapulids, kinorhynchs, loriciferans and nematomorphs. At le phylum des Cephalorhyncha qui inclut les priapulides, les the same time, the Palaeoscolecida share a number of char- kinorhynches, les loricifères et les nématomorphes. Cepen- acters with the lobopod-bearing Cambrian ecdysozoans, the dant, les Palaeoscolecida ont aussi quelques-uns des mêmes Xenusia. Xenusians commonly possess a terminal mouth, caractères que les Xenusia, ces écdysozaires cambriens qui a proboscis (although not retractable), and a thick integu- portaient des lobopodes.