Characterization and Modeling of the Human Left Atrium Using Optical Coherence Tomography
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Download PDF File
Folia Morphol. Vol. 78, No. 2, pp. 283–289 DOI: 10.5603/FM.a2018.0077 O R I G I N A L A R T I C L E Copyright © 2019 Via Medica ISSN 0015–5659 journals.viamedica.pl Observations of foetal heart veins draining directly into the left and right atria J.H. Kim1, O.H. Chai1, C.H. Song1, Z.W. Jin2, G. Murakami3, H. Abe4 1Department of Anatomy and Institute of Medical Sciences, Chonbuk National University Medical School, Jeonju, Korea 2Department of Anatomy, Wuxi Medical School, Jiangnan University, Wuxi, China 3Division of Internal Medicine, Jikou-kai Clinic of Home Visits, Sapporo, Japan 4Department of Anatomy, Akita University School of Medicine, Akita, Japan [Received: 19 June 2018; Accepted: 8 August 2018] Evaluation of semiserial sections of 14 normal hearts from human foetuses of gestational age 25–33 weeks showed that all of these hearts contained thin veins draining directly into the atria (maximum, 10 veins per heart). Of the 75 veins in these 14 hearts, 55 emptied into the right atrium and 20 into the left atrium. These veins were not accompanied by nerves, in contrast to tributaries of the great cardiac vein, and were negative for both smooth muscle actin (SMA) and CD34. However, the epithelium and venous wall of the anterior cardiac vein, the thickest of the direct draining veins, were strongly positive for SMA and CD34, respectively. In general, developing fibres in the vascular wall were positive for CD34, while the endothelium of the arteries and veins was strongly positive for the present DAKO antibody of SMA. -

Location of the Human Sinus Node in Black Africans
ogy: iol Cu ys r h re P n t & R y e s Anatomy & Physiology: Current m e o a t r a c n h A Research Meneas et al., Anat Physiol 2017, 7:5 ISSN: 2161-0940 DOI: 10.4172/2161-0940.1000279 Research article Open Access Location of the Human Sinus Node in Black Africans Meneas GC*, Yangni-Angate KH, Abro S and Adoubi KA Department of Cardiovascular and Thoracic Diseases, Bouake Teaching Hospital, Cote d’Ivoire, West-Africa *Corresponding author: Meneas GC, Department of Cardiovascular and Thoracic Diseases, Bouake Teaching Hospital, Cote d’Ivoire, West-Africa, Tel: +22507701532; E-mail: [email protected] Received Date: August 15, 2017; Accepted Date: August 22, 2017; Published Date: August 29, 2017 Copyright: © 2017 Meneas GC, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited. Abstract Objective: The purpose of this study was to describe, in 45 normal hearts of black Africans adults, the location of the sinoatrial node. Methods: After naked eye observation of the external epicardial area of the sinus node classically described as cavoatrial junction (CAJ), a histological study of the sinus node area was performed. Results: This study concluded that the sinus node is indistinguishable to the naked eye (97.77% of cases), but still identified histologically at the CAJ in the form of a cluster of nodal cells surrounded by abundant connective tissues. It is distinguished from the Myocardial Tissue. -

4B. the Heart (Cor) 1
Henry Gray (1821–1865). Anatomy of the Human Body. 1918. 4b. The Heart (Cor) 1 The heart is a hollow muscular organ of a somewhat conical form; it lies between the lungs in the middle mediastinum and is enclosed in the pericardium (Fig. 490). It is placed obliquely in the chest behind the body of the sternum and adjoining parts of the rib cartilages, and projects farther into the left than into the right half of the thoracic cavity, so that about one-third of it is situated on the right and two-thirds on the left of the median plane. Size.—The heart, in the adult, measures about 12 cm. in length, 8 to 9 cm. in breadth at the 2 broadest part, and 6 cm. in thickness. Its weight, in the male, varies from 280 to 340 grams; in the female, from 230 to 280 grams. The heart continues to increase in weight and size up to an advanced period of life; this increase is more marked in men than in women. Component Parts.—As has already been stated (page 497), the heart is subdivided by 3 septa into right and left halves, and a constriction subdivides each half of the organ into two cavities, the upper cavity being called the atrium, the lower the ventricle. The heart therefore consists of four chambers, viz., right and left atria, and right and left ventricles. The division of the heart into four cavities is indicated on its surface by grooves. The atria 4 are separated from the ventricles by the coronary sulcus (auriculoventricular groove); this contains the trunks of the nutrient vessels of the heart, and is deficient in front, where it is crossed by the root of the pulmonary artery. -

Sudden Death in Racehorses: Postmortem Examination Protocol
VDIXXX10.1177/1040638716687004Sudden death in racehorsesDiab et al. 687004research-article2017 Special Issue Journal of Veterinary Diagnostic Investigation 1 –8 Sudden death in racehorses: postmortem © 2017 The Author(s) Reprints and permissions: sagepub.com/journalsPermissions.nav examination protocol DOI: 10.1177/1040638716687004 jvdi.sagepub.com Santiago S. Diab,1 Robert Poppenga, Francisco A. Uzal Abstract. In racehorses, sudden death (SD) associated with exercise poses a serious risk to jockeys and adversely affects racehorse welfare and the public perception of horse racing. In a majority of cases of exercise-associated sudden death (EASD), there are no gross lesions to explain the cause of death, and an examination of the cardiovascular system and a toxicologic screen are warranted. Cases of EASD without gross lesions are often presumed to be sudden cardiac deaths (SCD). We describe an equine SD autopsy protocol, with emphasis on histologic examination of the heart (“cardiac histology protocol”) and a description of the toxicologic screen performed in racehorses in California. By consistently utilizing this standardized autopsy and cardiac histology protocol, the results and conclusions from postmortem examinations will be easier to compare within and across institutions over time. The generation of consistent, reliable, and comparable multi-institutional data is essential to improving the understanding of the cause(s) and pathogenesis of equine SD, including EASD and SCD. Key words: Cardiac autopsy; equine; exercise; racehorses; -

Ultrastructure of the Normal Atrial Endocardium R
Brit. Heart J., 1966, 28, 785. Ultrastructure of the Normal Atrial Endocardium R. A. LANNIGAN AND SALEH A. ZAKI From the Department of Pathology, University of Birmingham, and the Department of Pathology, University of Assiut, U.A.R. Surgical biopsies of the heart and early needle Thin sections were then cut and examined on an A.E.I. "biopsies" at necropsy now permit ultrastructural E M 6 electron microscope. investigations into many cardiac conditions. There For light microscopy, blocks of the whole circum- are, however, very few such studies on normal human ference were embedded in paraffin and sections were stained with Ehrlich's hTmatoxylin and eosin, van myocardium and none are available on human or Gieson's stain, Lawson's modification of the Weigert- animal endocardium. The commonest surgical Sheridan elastic stain, the periodic-acid Schiff reaction, specimens from the heart are from the left or right and Hale's dialysed iron. atrial appendages, and this material has been used to study normal structure, as a baseline for the investigation of cardiac diseases such as rheumatic RESULTS heart disease and fibro-elastosis, etc. For obvious Light Microscopy reasons, it is difficult to obtain fresh material from "normal" hearts, and specimens were selected for The structure of the left atrial wall has been electron microscopy which were within normal described by Koniger (1903), Nagayo (1909), Von limits on the light microscope (Lannigan, 1959). Glahn (1926), and Gross (1935), and the structure of the atrial appendages, which was shown to be similar to that of the atrium, was described by MATERIAL AND METHODS Waaler (1952), Lannigan (1959), and Ling, Wang, Three left atrial appendages and three right append- and Kuo (1959). -

Abnormally Enlarged Singular Thebesian Vein in Right Atrium
Open Access Case Report DOI: 10.7759/cureus.16300 Abnormally Enlarged Singular Thebesian Vein in Right Atrium Dilip Kumar 1 , Amit Malviya 2 , Bishwajeet Saikia 3 , Bhupen Barman 4 , Anunay Gupta 5 1. Cardiology, Medica Institute of Cardiac Sciences, Kolkata, IND 2. Cardiology, North Eastern Indira Gandhi Regional Institute of Health and Medical Sciences, Shillong, IND 3. Anatomy, North Eastern Indira Gandhi Regional Institute of Health and Medical Sciences, Shillong, IND 4. Internal Medicine, North Eastern Indira Gandhi Regional Institute of Health and Medical Sciences, Shillong, IND 5. Cardiology, Vardhman Mahavir Medical College (VMMC) and Safdarjung Hospital, New Delhi, IND Corresponding author: Amit Malviya, [email protected] Abstract Thebesian veins in the heart are subendocardial venoluminal channels and are usually less than 0.5 mm in diameter. The system of TV either opens a venous (venoluminal) or an arterial (arterioluminal) channel directly into the lumen of the cardiac chambers or via some intervening spaces (venosinusoidal/ arteriosinusoidal) termed as sinusoids. Enlarged thebesian veins are reported in patients with congenital heart disease and usually, multiple veins are enlarged. Very few reports of such abnormal enlargement are there in the absence of congenital heart disease, but in all such cases, they are multiple and in association with coronary artery microfistule. We report a very rare case of a singular thebesian vein in the right atrium, which was abnormally enlarged. It is important to recognize because it can be confused with other cardiac structures like coronary sinus during diagnostic or therapeutic catheterization and can lead to cardiac injury and complications if it is attempted to cannulate it or pass the guidewires. -

Anatomy of the Heart
Anatomy of the Heart DR. SAEED VOHRA DR. SANAA AL-SHAARAWI OBJECTIVES • At the end of the lecture, the student should be able to : • Describe the shape of heart regarding : apex, base, sternocostal and diaphragmatic surfaces. • Describe the interior of heart chambers : right atrium, right ventricle, left atrium and left ventricle. • List the orifices of the heart : • Right atrioventricular (Tricuspid) orifice. • Pulmonary orifice. • Left atrioventricular (Mitral) orifice. • Aortic orifice. • Describe the innervation of the heart • Briefly describe the conduction system of the Heart The Heart • It lies in the middle mediastinum. • It is surrounded by a fibroserous sac called pericardium which is differentiated into an outer fibrous layer (Fibrous pericardium) & inner serous sac (Serous pericardium). • The Heart is somewhat pyramidal in shape, having: • Apex • Sterno-costal (anterior surface) • Base (posterior surface). • Diaphragmatic (inferior surface) • It consists of 4 chambers, 2 atria (right& left) & 2 ventricles (right& left) Apex of the heart • Directed downwards, forwards and to the left. • It is formed by the left ventricle. • Lies at the level of left 5th intercostal space 3.5 inch from midline. Note that the base of the heart is called the base because the heart is pyramid shaped; the base lies opposite the apex. The heart does not rest on its base; it rests on its diaphragmatic (inferior) surface Sterno-costal (anterior)surface • Divided by coronary (atrio- This surface is formed mainly ventricular) groove into : by the right atrium and the right . Atrial part, formed mainly by ventricle right atrium. Ventricular part , the right 2/3 is formed by right ventricle, while the left l1/3 is formed by left ventricle. -

The American Society of Echocardiography
1 THE AMERICAN SOCIETY OF ECHOCARDIOGRAPHY RECOMMENDATIONS FOR CARDIAC CHAMBER QUANTIFICATION IN ADULTS: A QUICK REFERENCE GUIDE FROM THE ASE WORKFLOW AND LAB MANAGEMENT TASK FORCE Accurate and reproducible assessment of cardiac chamber size and function is essential for clinical care. A standardized methodology creates a common approach to the assessment of cardiac structure and function both within and between echocardiography labs. This facilitates better communication and improves the ability to compare results between studies as well as differentiate normal from abnormal findings in an individual patient. This document summarizes key points from the 2015 ASE Chamber Quantification Guideline and is meant to serve as quick reference for sonographers and interpreting physicians. It is designed to provide guidance on chamber quantification for adult patients; a separate ASE Guidelines document that details recommended quantification methods in the pediatric age group has also been published and should be used for patients <18 years of age (3). (1) For details of the methodology and the rationale for current recommendations, the interested reader is referred to the complete Guideline statement. Figures and tables are reproduced from ASE Guidelines. (1,2) Table of Contents: 1. Left Ventricle (LV) Size and Function p. 2 a. LV Size p. 2 i. Linear Measurements p. 2 ii. Volume Measurements p. 2 iii. LV Mass Calculations p. 3 b. Left Ventricular Function Assessment p. 4 i. Global Systolic Function Parameters p. 4 ii. Regional Function p. 5 2. Right Ventricle (RV) Size and Function p. 6 a. RV Size p. 6 b. RV Function p. 8 3. Atria p. -

The Biomechanical Puncture Study of the Fossa Ovalis in Human and Porcine Hearts Daniel S
The Biomechanical Puncture Study of the Fossa Ovalis in Human and Porcine Hearts Daniel S. Balto, Steven A. Howard, BMEn., Paul A. Iaizzo, Ph.D.2 Departments of Biomedical Engineering1 and Surgery2 University of Minnesota, Minneapolis, MN 55455 Introduction Apparatus Conclusion Approximately 1 in 4 humans on this earth is born with a congenital heart •The apparatus designed for this study was great for initial testing of the defect that involves the failure of the complete formation of the fossa ovalis tissue properties of the fossa ovalis of large mammalian hearts, either of membrane between the left and right atria1. This defect, better known as a PFO research animals or of human donor hearts. However, physiological or Patent Fossa Ovalis is an opening in the fossa ovalis membrane that results properties weren’t really observed in the testing and so that is a starting from the failure of the fetal structures (septum primum and septum secundum) point for advancing the quality of this research. The ability for the system to from closing together at the moment of birth2. The opening can be classified in have interchangeable depression, puncture, and dilation devices adds to the a variety of ways, but the most common are “shunts” which cause blood to flexibility of the current setup. The visualization of the inside of the heart flow from one atria to the other. A shunt that results in the blood flowing from adds a new dimension to this research as well since it is able to visualize all the left to right atria will cause the blood pressure to increase in the right side cardiac structures under ~10 mmHg of pressure. -

Structures of the Heart
Anatomy Tip Structures of the Heart In an effort to aid Health Information Management Coding Professionals for ICD-10, the following anatomy tip is provided with an educational intent. TIP: The Heart is made up of three main structures: 1. The Pericardium 2. The Heart Wall 3. The Chambers of the Heart The pericardium surrounds the heart. The outer layer, called the fibrous pericardium, secures the heart to surrounding structures like the blood vessels and the diaphragm. The inner layer, called the serous pericardium, is a double-layered section of the heart. Inside the two layers, serous fluid, known as pericardial fluid, lubricates and helps the heart move fluidly when beating. The heart wall is made up of three tissue layers: the epicardium, the myocardium, and the endocardium. The epicardium, which is the outermost layer, is also known as the visceral pericardium because it is also the inner wall of the pericardium. The middle layer, known as the myocardium, is formed out of contracting muscle. The endocardium, the innermost layer, covers heart valves and acts as a lining of the heart chambers. It is also in contact with the blood that is pumped through the heart, in order to push blood into the lungs and throughout the rest of the body. The four heart chambers are crucial to the heart’s function, composed of the atria, the right atrium and left atrium, and ventricles, the right ventricle and left ventricle. The right and left atria are thin-walled chambers responsible for receiving blood from veins, while the left and right ventricle are thick-walled chambers responsible for pumping blood out of the heart. -

Premature Obliteration of the Foramen Ovale by G
PREMATURE OBLITERATION OF THE FORAMEN OVALE BY G. AUSTIN GRESHAM From the Department of Pathology, University of Cambridge Received August 16, 1955 Obliteration of the foramen ovale occurring during intrauterine life is a rare condition. It throws some light on the mechanism of development of endocardial fibroelastosis and also upon the factors concerned in determining the size of the aorta and of the cardiac chambers. CASE HISTORY The patient was the second child of a mother (aet. 21) whose previous obstetric history was normal. Apart from two periods of rather rapid gain of weight for which no cause could be found, the pregnancy was uneventful. The child was eleven days postmature; labour was induced with an enema and lasted forty-five minutes. The infant cried lustily but was cyanosed: the heart was clinically normal. Abnormalities were present in all four limbs. The left radius and ulna were absent and rudimentary digits were present on the skin over the distal end of the limb. Terminal phalanges were absent in the fingers of the right hand, and four toes were present on each foot with a rudimentary fifth digit on the left foot. Cyanosis and dyspniea became more intense and the child died three hours after birth despite the use of oxygen. NECROPSY The body was that of a full-term male infant (weight 3430 g.). The limbs were abnormal as previously described. The lips were blue-black in colour. On the septal wall of the right atrium a hemispherical grey-white area (10x 12 x 4 mm. deep) with a central dimple filled in the usual site of the fossa ovalis (Fig. -
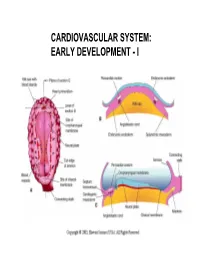
Cardiovascular System: Early Development
CARDIOVASCULAR SYSTEM: EARLY DEVELOPMENT - I CARDIOVASCULAR SYSTEM: EARLY DEVELOPMENT - II HEART AND ITS NEIGHBORHOOD- I HEART AND ITS NEIGHBORHOOD- II BLOOD VESSELS OF THE EMBRYO (at 26 days) FROM TUBE TO FOUR CHAMBERS EXTERNAL VIEW NORMAL : Loop to the RIGHT: Levocardia! ABNORMAL: Loop to the LEFT: Dextrocardia! FROM TUBE TO FOUR CHAMBERS INTERNAL VIEW FOUR CHAMBERS- ULTRASOUND VIEW @ 20 wks ATRIAL SEPTUM FORMATION- I ATRIAL SEPTUM FORMATION- II ATRIAL SEPTUM FORMATION- III ATRIAL SEPTUM FORMATION- IV ATRIAL SEPTUM FORMATION- V THE DEFINITIVE RIGHT ATRIUM **NOTE: In 25% of normal population, the foramen ovale remains ‘probe patent’. THE DEFINITIVE LEFT ATRIUM ATRIAL SEPTAL DEFECTS- “Fossa Ovalis” type ATRIAL SEPTAL DEFECTS- Unrelated to Foramen Ovale AORTIC ARCHES AND DERIVATIVES - I AORTIC ARCHES AND DERIVATIVES - II RECURRENT LARYNGEAL NERVES Right vs Left PATENT DUCTUS ARTERIOSUS Prostaglandin: Keeps the duct Patent Indomethacin: Closes the duct. RIGHT AORTIC ARCH: Mirror image branching ABERRANT RIGHT SUBCLAVIAN ARTERY: Occurs in 0.5% of people. Usually asymptomatic. DOUBLE AORTIC ARCH: “Vascular ring” Causes airway obstruction, stridor in infancy. COARCTATION OF THE AORTA THE CARDINAL VEINS AND THE VENAE CAVAE THE CARDINAL VEINS AND THE VENAE CAVAE SINUS VENOSUS AND THE CORONARY SINUS PERSISTENT LEFT SVC 0.3% of general population. 4 % of patients with Cong. Ht Dis. Usually drains to Coronary sinus. Usually asymptomatic. Enlarged coronary sinus is a clue. Left SVC to coronary sinus UMBILICAL AND VITELLINE VEINS- I: Liver, portal vein and ductus venosus. © 2005 Elsevier UMBILICAL AND VITELLINE VEINS- II: Liver, portal vein and ductus venosus. LYMPHATIC SYSTEM- I LYMPHATIC SYSTEM- II FETAL CIRCULATION POSTNATAL CIRCULATION.