Downloaded at Panorama Public ( Email: [email protected] , Password: Jv7z4#GC)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Peking University-Juntendo University Joint Symposium on Cancer Research and Treatment ADAM28 (A Disintegrin and Metalloproteinase 28) in Cancer Cell Proliferation and Progression
Whatʼs New from Juntendo University, Tokyo Juntendo Medical Journal 2017. 63(5), 322-325 Peking University - Juntendo University Joint Symposium on Cancer Research and Treatment ADAM28 (a Disintegrin and Metalloproteinase 28) in Cancer Cell Proliferation and Progression YASUNORI OKADA* *Department of Pathophysiology for Locomotive and Neoplastic Diseases, Juntendo University Graduate School of Medicine, Tokyo, Japan A disintegrinandmetalloproteinase 28 (ADAM28) is overexpressedpredominantlyby carcinoma cells in more than 70% of the non-small cell lung carcinomas, showing positive correlations with carcinoma cell proliferation and metastasis. ADAM28 cleaves insulin-like growth factor binding protein-3 (IGFBP-3) in the IGF-I/IGFBP-3 complex, leading to stimulation of cell proliferation by intact IGF-I released from the complex. ADAM28 also degrades von Willebrand factor (VWF), which induces apoptosis in human carcinoma cell lines with negligible ADAM28 expression, andthe VWF digestionby ADAM28-expressing carcinoma cells facilitates them to escape from VWF-induced apoptosis, resulting in promotion of metastasis. We have developed human antibodies against ADAM28 andshown that one of them significantly inhibits tumor growth andmetastasis using lung adenocarcinoma cells. Our data suggest that ADAM28 may be a new molecular target for therapy of the patients with ADAM28-expressing non-small cell lung carcinoma. Key words: a disintegrin and metalloproteinase 28 (ADAM28), cell proliferation, invasion, metastasis, human antibody inhibitor Introduction human cancers 2). However, development of the synthetic inhibitors of MMPs andtheir application Cancer cell proliferation andprogression are for treatment of the cancer patients failed 3). modulated by proteolytic cleavage of tissue micro- On the other hand, members of the ADAM (a environmental factors such as extracellular matrix disintegrin and metalloproteinase) gene family, (ECM), growth factors andcytokines, receptors another family belonging to the metzincin gene andcell adhesionmolecules. -

Genome-Wide Analysis Reveals Selection Signatures Involved in Meat Traits and Local Adaptation in Semi-Feral Maremmana Cattle
Genome-Wide Analysis Reveals Selection Signatures Involved in Meat Traits and Local Adaptation in Semi-Feral Maremmana Cattle Slim Ben-Jemaa, Gabriele Senczuk, Elena Ciani, Roberta Ciampolini, Gennaro Catillo, Mekki Boussaha, Fabio Pilla, Baldassare Portolano, Salvatore Mastrangelo To cite this version: Slim Ben-Jemaa, Gabriele Senczuk, Elena Ciani, Roberta Ciampolini, Gennaro Catillo, et al.. Genome-Wide Analysis Reveals Selection Signatures Involved in Meat Traits and Local Adaptation in Semi-Feral Maremmana Cattle. Frontiers in Genetics, Frontiers, 2021, 10.3389/fgene.2021.675569. hal-03210766 HAL Id: hal-03210766 https://hal.inrae.fr/hal-03210766 Submitted on 28 Apr 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License ORIGINAL RESEARCH published: 28 April 2021 doi: 10.3389/fgene.2021.675569 Genome-Wide Analysis Reveals Selection Signatures Involved in Meat Traits and Local Adaptation in Semi-Feral Maremmana Cattle Slim Ben-Jemaa 1, Gabriele Senczuk 2, Elena Ciani 3, Roberta -

Seq2pathway Vignette
seq2pathway Vignette Bin Wang, Xinan Holly Yang, Arjun Kinstlick May 19, 2021 Contents 1 Abstract 1 2 Package Installation 2 3 runseq2pathway 2 4 Two main functions 3 4.1 seq2gene . .3 4.1.1 seq2gene flowchart . .3 4.1.2 runseq2gene inputs/parameters . .5 4.1.3 runseq2gene outputs . .8 4.2 gene2pathway . 10 4.2.1 gene2pathway flowchart . 11 4.2.2 gene2pathway test inputs/parameters . 11 4.2.3 gene2pathway test outputs . 12 5 Examples 13 5.1 ChIP-seq data analysis . 13 5.1.1 Map ChIP-seq enriched peaks to genes using runseq2gene .................... 13 5.1.2 Discover enriched GO terms using gene2pathway_test with gene scores . 15 5.1.3 Discover enriched GO terms using Fisher's Exact test without gene scores . 17 5.1.4 Add description for genes . 20 5.2 RNA-seq data analysis . 20 6 R environment session 23 1 Abstract Seq2pathway is a novel computational tool to analyze functional gene-sets (including signaling pathways) using variable next-generation sequencing data[1]. Integral to this tool are the \seq2gene" and \gene2pathway" components in series that infer a quantitative pathway-level profile for each sample. The seq2gene function assigns phenotype-associated significance of genomic regions to gene-level scores, where the significance could be p-values of SNPs or point mutations, protein-binding affinity, or transcriptional expression level. The seq2gene function has the feasibility to assign non-exon regions to a range of neighboring genes besides the nearest one, thus facilitating the study of functional non-coding elements[2]. Then the gene2pathway summarizes gene-level measurements to pathway-level scores, comparing the quantity of significance for gene members within a pathway with those outside a pathway. -

To Study Mutant P53 Gain of Function, Various Tumor-Derived P53 Mutants
Differential effects of mutant TAp63γ on transactivation of p53 and/or p63 responsive genes and their effects on global gene expression. A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science By Shama K Khokhar M.Sc., Bilaspur University, 2004 B.Sc., Bhopal University, 2002 2007 1 COPYRIGHT SHAMA K KHOKHAR 2007 2 WRIGHT STATE UNIVERSITY SCHOOL OF GRADUATE STUDIES Date of Defense: 12-03-07 I HEREBY RECOMMEND THAT THE THESIS PREPARED UNDER MY SUPERVISION BY SHAMA KHAN KHOKHAR ENTITLED Differential effects of mutant TAp63γ on transactivation of p53 and/or p63 responsive genes and their effects on global gene expression BE ACCEPTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF Master of Science Madhavi P. Kadakia, Ph.D. Thesis Director Daniel Organisciak , Ph.D. Department Chair Committee on Final Examination Madhavi P. Kadakia, Ph.D. Steven J. Berberich, Ph.D. Michael Leffak, Ph.D. Joseph F. Thomas, Jr., Ph.D. Dean, School of Graduate Studies 3 Abstract Khokhar, Shama K. M.S., Department of Biochemistry and Molecular Biology, Wright State University, 2007 Differential effect of TAp63γ mutants on transactivation of p53 and/or p63 responsive genes and their effects on global gene expression. p63, a member of the p53 gene family, known to play a role in development, has more recently also been implicated in cancer progression. Mice lacking p63 exhibit severe developmental defects such as limb truncations, abnormal skin, and absence of hair follicles, teeth, and mammary glands. Germline missense mutations of p63 have been shown to be responsible for several human developmental syndromes including SHFM, EEC and ADULT syndromes and are associated with anomalies in the development of organs of epithelial origin. -
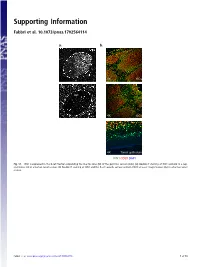
Supporting Information
Supporting Information Fabbri et al. 10.1073/pnas.1702564114 a. b. GC 4X GCs 20x GC 4X GCs 4X Tonsil epithelium ICN1 CD20 DAPI Fig. S1. ICN1 is expressed in the B-cell fraction populating the mantle zone (M) of the germinal centers (GCs). (A) Double IF staining of ICN1 and AID in a rep- resentative GC in a human tonsil section. (B) Double IF staining of ICN1 and the B-cell–specific surface antigen CD20 at lower magnification (4×) in a human tonsil section. Fabbri et al. www.pnas.org/cgi/content/short/1702564114 1of10 Fig. S2. ICN1 expression analysis in a panel of primary CLL cases and PBMC. (A) IB analysis of ICN1 and control β-actin in a panel of 124 CLL PB primary CLL cases, (B) in primary NOTCH1–wild-type CLL cells treated with the γ-secretase inhibitor Compound E (CpE, 500 nM, 8 h) or control DMSO, and (C) in PBMC protein extracts and representative primary CLL cases expressing ICN1. Samples are color-coded based on the NOTCH1 mutational status [red, clonal NOTCH1 PEST-truncating events; orange, subclonal NOTCH1 PEST-truncating events; blue, RAG-mediated NOTCH1 translocation (83); and black, NOTCH1–wild-type]. Samples in gray were excluded from the analysis because of low quality of the protein lysate, low viability, or low leukemic representation. Color-coded arrows indicate cases subjected to RNA-Seq analysis: dark red denotes NOTCH1-mutated cases expressing ICN1; blue, NOTCH1–wild-type cases expressing ICN1; and green, ICN1− NOTCH1–wild-type cases. Abbreviations: MO+DL1, MO1043 cells cocultured on OP9-DL1 cells (54); s.e., short exposure; l.e., long exposure. -

Pancancer Progression Human Vjune2017
Gene Symbol Accession Alias/Prev Symbol Official Full Name AAMP NM_001087.3 - angio-associated, migratory cell protein ABI3BP NM_015429.3 NESHBP|TARSH ABI family, member 3 (NESH) binding protein ACHE NM_000665.3 ACEE|ARACHE|N-ACHE|YT acetylcholinesterase ACTG2 NM_001615.3 ACT|ACTA3|ACTE|ACTL3|ACTSG actin, gamma 2, smooth muscle, enteric ACVR1 NM_001105.2 ACTRI|ACVR1A|ACVRLK2|ALK2|FOP|SKR1|TSRI activin A receptor, type I ACVR1C NM_145259.2 ACVRLK7|ALK7 activin A receptor, type IC ACVRL1 NM_000020.1 ACVRLK1|ALK-1|ALK1|HHT|HHT2|ORW2|SKR3|TSR-I activin A receptor type II-like 1 ADAM15 NM_207195.1 MDC15 ADAM metallopeptidase domain 15 ADAM17 NM_003183.4 ADAM18|CD156B|CSVP|NISBD|TACE ADAM metallopeptidase domain 17 ADAM28 NM_014265.4 ADAM 28|ADAM23|MDC-L|MDC-Lm|MDC-Ls|MDCL|eMDC II|eMDCII ADAM metallopeptidase domain 28 ADAM8 NM_001109.4 CD156|MS2 ADAM metallopeptidase domain 8 ADAM9 NM_001005845.1 CORD9|MCMP|MDC9|Mltng ADAM metallopeptidase domain 9 ADAMTS1 NM_006988.3 C3-C5|METH1 ADAM metallopeptidase with thrombospondin type 1 motif, 1 ADAMTS12 NM_030955.2 PRO4389 ADAM metallopeptidase with thrombospondin type 1 motif, 12 ADAMTS8 NM_007037.4 ADAM-TS8|METH2 ADAM metallopeptidase with thrombospondin type 1 motif, 8 ADAP1 NM_006869.2 CENTA1|GCS1L|p42IP4 ArfGAP with dual PH domains 1 ADD1 NM_001119.4 ADDA adducin 1 (alpha) ADM2 NM_001253845.1 AM2|dJ579N16.4 adrenomedullin 2 ADRA2B NM_000682.4 ADRA2L1|ADRA2RL1|ADRARL1|ALPHA2BAR|alpha-2BAR adrenoceptor alpha 2B AEBP1 NM_001129.3 ACLP AE binding protein 1 AGGF1 NM_018046.3 GPATC7|GPATCH7|HSU84971|HUS84971|VG5Q -

IBRAHIM ALDEAILEJ Phd.2013.Pdf
Bangor University DOCTOR OF PHILOSOPHY Identification and functional characterisation of germ line genes in human cancer cells Aldeailej, Ibrahim Award date: 2013 Awarding institution: Bangor University Link to publication General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal ? Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Download date: 09. Oct. 2021 Bangor University School of Biological Sciences Identification and functional characterisation of germ line genes in human cancer cells Ph.D. Thesis 2013 IBRAHIM ALDEAILEJ Declaration and Consent Details of the Work I hereby agree to deposit the following item in the digital repository maintained by Bangor University and/or in any other repository authorized for use by Bangor University. Author Name: IBRAHIM MOHAMMED ALDEAILEJ Title: Identification and functional characterisation of germ line genes in human cancer cells Supervisor/Department: Dr. Ramsay James McFarlane/ School of Biological Sciences Funding body (if any): kingdom of Saudi Arabia Government Qualification/Degree obtained: Ph.D. -

Supp Table 6.Pdf
Supplementary Table 6. Processes associated to the 2037 SCL candidate target genes ID Symbol Entrez Gene Name Process NM_178114 AMIGO2 adhesion molecule with Ig-like domain 2 adhesion NM_033474 ARVCF armadillo repeat gene deletes in velocardiofacial syndrome adhesion NM_027060 BTBD9 BTB (POZ) domain containing 9 adhesion NM_001039149 CD226 CD226 molecule adhesion NM_010581 CD47 CD47 molecule adhesion NM_023370 CDH23 cadherin-like 23 adhesion NM_207298 CERCAM cerebral endothelial cell adhesion molecule adhesion NM_021719 CLDN15 claudin 15 adhesion NM_009902 CLDN3 claudin 3 adhesion NM_008779 CNTN3 contactin 3 (plasmacytoma associated) adhesion NM_015734 COL5A1 collagen, type V, alpha 1 adhesion NM_007803 CTTN cortactin adhesion NM_009142 CX3CL1 chemokine (C-X3-C motif) ligand 1 adhesion NM_031174 DSCAM Down syndrome cell adhesion molecule adhesion NM_145158 EMILIN2 elastin microfibril interfacer 2 adhesion NM_001081286 FAT1 FAT tumor suppressor homolog 1 (Drosophila) adhesion NM_001080814 FAT3 FAT tumor suppressor homolog 3 (Drosophila) adhesion NM_153795 FERMT3 fermitin family homolog 3 (Drosophila) adhesion NM_010494 ICAM2 intercellular adhesion molecule 2 adhesion NM_023892 ICAM4 (includes EG:3386) intercellular adhesion molecule 4 (Landsteiner-Wiener blood group)adhesion NM_001001979 MEGF10 multiple EGF-like-domains 10 adhesion NM_172522 MEGF11 multiple EGF-like-domains 11 adhesion NM_010739 MUC13 mucin 13, cell surface associated adhesion NM_013610 NINJ1 ninjurin 1 adhesion NM_016718 NINJ2 ninjurin 2 adhesion NM_172932 NLGN3 neuroligin -
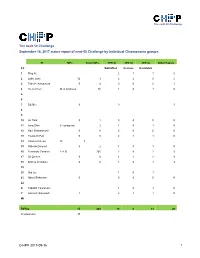
Next-MP50 Status Report
The neXt-50 Challenge The neXt-50 Challenge September 16, 2017 status report of next-50 Challenge by individual Chromosome groups. PI MPs Silver MPs JPR SI JPR SI JPR SI Other Papers Ch Submitted In press In revision 1 Ping Xu 2 1 1 0 2 Lydie Lane 12 3 2 2 0 2 3 Takeshi Kawamura 0 4 0 0 0 1 4 Yu-Ju Chen 26 in progress 86 1 0 1 0 5 6 7 Ed Nice 0 0 1 8 9 10 Jin Park 0 1 0 0 0 0 11 Jong Shin 2 in progress 2 1 0 1 0 12 Ravi Sirdeshmukh 0 0 0 0 0 0 13 Young-Ki Paik 0 5 2 1 1 0 14 CharLes Pineau 12 3 15 GiLberto Domont 0 2 1 0 1 0 16 Fernando CorraLes 2 (+ 9) 165 2 0 2 5 17 GiL Omenn 0 0 3 1 2 8 18 Andrey Archakov 0 0 1 0 1 3 19 20 Siqi Liu 1 0 1 21 Albert Sickmann 0 0 0 0 0 22 X Tadashi Yamamoto 1 0 1 0 Y Hosseini SaLekdeh 1 2 1 1 0 Mt TOTAL 15 268 19 6 13 20 (in progress) 37 C-HPP 2017-09-16 1 The neXt-50 Challenge Chromosome 1 (Ping Xu) PIC Leaders: Ping Xu, Fuchu He Contributing labs: Ping Xu, Beijing Proteome Research Center Fuchu He, Beijing Proteome Research Center Dong Yang, Beijing Proteome Research Center Wantao Ying, Beijing Proteome Research Center Pengyuan Yang, Fudan University Siqi Liu, Beijing Genome Institute Qinyu He, Jinan University Major lab members or partners contributing to the neXt50: Yao Zhang (Beijing Proteome Research Center), Yihao Wang (Beijing Proteome Research Center), Cuitong He (Beijing Proteome Research Center), Wei Wei (Beijing Proteome Research Center), Yanchang Li (Beijing Proteome Research Center), Feng Xu (Beijing Proteome Research Center), Xuehui Peng (Beijing Proteome Research Center). -

Ultra-Processed Food Targets Bone Quality Via Endochondral Ossification
Bone Research www.nature.com/boneres ARTICLE OPEN Ultra-processed food targets bone quality via endochondral ossification Janna Zaretsky1, Shelley Griess-Fishheimer1, Adi Carmi1, Tamara Travinsky Shmul1, Lior Ofer2, Tali Sinai1, Svetlana Penn1, Ron Shahar2 and Efrat Monsonego-Ornan 1 Ultra-processed foods have known negative implications for health; however, their effect on skeletal development has never been explored. Here, we show that young rats fed ultra-processed food rich in fat and sugar suffer from growth retardation due to lesions in their tibial growth plates. The bone mineral density decreases significantly, and the structural parameters of the bone deteriorate, presenting a sieve-like appearance in the cortices and poor trabecular parameters in long bones and vertebrae. This results in inferior mechanical performance of the entire bone with a high fracture risk. RNA sequence analysis of the growth plates demonstrated an imbalance in extracellular matrix formation and degradation and impairment of proliferation, differentiation and mineralization processes. Our findings highlight, for the first time, the severe impact of consuming ultra-processed foods on the growing skeleton. This pathology extends far beyond that explained by the known metabolic effects, highlighting bone as a new target for studies of modern diets. Bone Research (2021) ;9:14 https://doi.org/10.1038/s41413-020-00127-9 1234567890();,: INTRODUCTION penetration of blood vessels and bone tissue formation by The vertebrate skeleton has evolved as a dynamic system that osteoblasts.1,2,4,6 The EO process is tightly orchestrated by various serves numerous functions, such as protecting internal organs, signaling molecules and transcription factors, including transform- creating attachment sites for muscles to produce locomotion, ing growth factor β/bone morphogenetic protein (BMP), fibroblast providing a reservoir for minerals, and serving as a hematopoietic growth factor (FGF), Wnt, hedgehog, and the transcription factor niche. -

Overexpression of the Proneural Transcription Factor ASCL1 In
Malli et al. Molecular Cytogenetics (2018) 11:3 DOI 10.1186/s13039-018-0355-7 CASE REPORT Open Access Overexpression of the proneural transcription factor ASCL1 in chronic lymphocytic leukemia with a t(12;14)(q23.2;q32.3) Theodora Malli1, Melanie Rammer1, Sabrina Haslinger1, Jonathan Burghofer1, Sonja Burgstaller2, Hans-Christian Boesmueller3,4, Renate Marschon1, Wolfgang Kranewitter1, Martin Erdel1, Sabine Deutschbauer1 and Gerald Webersinke1* Abstract Background: Translocations of the IGH locus on 14q32.3 are present in about 8% of patients with chronic lymphocytic leukemia (CLL) and contribute to leukemogenesis by deregulating the expression of the IGH-partner genes. Identification of these genes and investigation of the downstream effects of their deregulation can reveal disease-causing mechanisms. Case presentation: We report on the molecular characterization of a novel t(12;14)(q23.2;q32.3) in CLL. As a consequence of the rearrangement ASCL1 was brought into proximity of the IGHJ-Cμ enhancer and was highly overexpressed in the aberrant B-cells of the patient, as shown by qPCR and immunohistochemistry. ASCL1 encodes for a transcription factor acting as a master regulator of neurogenesis, is overexpressed in neuroendocrine tumors and a promising therapeutic target in small cell lung cancer (SCLC). Its overexpression has also been recently reported in acute adult T-cell leukemia/lymphoma. To examine possible downstream effects of the ASCL1 upregulation in CLL, we compared the gene expression of sorted CD5+ cells of the translocation patient to that of CD19+ B-cells from seven healthy donors and detected 176 significantly deregulated genes (Fold Change ≥2, FDR p ≤ 0.01). -

Novel Targets of Apparently Idiopathic Male Infertility
International Journal of Molecular Sciences Review Molecular Biology of Spermatogenesis: Novel Targets of Apparently Idiopathic Male Infertility Rossella Cannarella * , Rosita A. Condorelli , Laura M. Mongioì, Sandro La Vignera * and Aldo E. Calogero Department of Clinical and Experimental Medicine, University of Catania, 95123 Catania, Italy; [email protected] (R.A.C.); [email protected] (L.M.M.); [email protected] (A.E.C.) * Correspondence: [email protected] (R.C.); [email protected] (S.L.V.) Received: 8 February 2020; Accepted: 2 March 2020; Published: 3 March 2020 Abstract: Male infertility affects half of infertile couples and, currently, a relevant percentage of cases of male infertility is considered as idiopathic. Although the male contribution to human fertilization has traditionally been restricted to sperm DNA, current evidence suggest that a relevant number of sperm transcripts and proteins are involved in acrosome reactions, sperm-oocyte fusion and, once released into the oocyte, embryo growth and development. The aim of this review is to provide updated and comprehensive insight into the molecular biology of spermatogenesis, including evidence on spermatogenetic failure and underlining the role of the sperm-carried molecular factors involved in oocyte fertilization and embryo growth. This represents the first step in the identification of new possible diagnostic and, possibly, therapeutic markers in the field of apparently idiopathic male infertility. Keywords: spermatogenetic failure; embryo growth; male infertility; spermatogenesis; recurrent pregnancy loss; sperm proteome; DNA fragmentation; sperm transcriptome 1. Introduction Infertility is a widespread condition in industrialized countries, affecting up to 15% of couples of childbearing age [1]. It is defined as the inability to achieve conception after 1–2 years of unprotected sexual intercourse [2].