Specificity and Mechanism of the Histone Methyltransferase Pr-Set7
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Plant SET Domain-Containing Proteins: Structure, Function and Regulation
Biochimica et Biophysica Acta 1769 (2007) 316–329 www.elsevier.com/locate/bbaexp Review Plant SET domain-containing proteins: Structure, function and regulation Danny W-K Ng, Tao Wang, Mahesh B. Chandrasekharan 1, Rodolfo Aramayo, ⁎ Sunee Kertbundit 2, Timothy C. Hall Institute of Developmental and Molecular Biology and Department of Biology, Texas A&M University, College Station, TX 77843-3155, USA Received 27 October 2006; received in revised form 3 April 2007; accepted 4 April 2007 Available online 12 April 2007 Abstract Modification of the histone proteins that form the core around which chromosomal DNA is looped has profound epigenetic effects on the accessibility of the associated DNA for transcription, replication and repair. The SET domain is now recognized as generally having methyltransferase activity targeted to specific lysine residues of histone H3 or H4. There is considerable sequence conservation within the SET domain and within its flanking regions. Previous reviews have shown that SET proteins from Arabidopsis and maize fall into five classes according to their sequence and domain architectures. These classes generally reflect specificity for a particular substrate. SET proteins from rice were found to fall into similar groupings, strengthening the merit of the approach taken. Two additional classes, VI and VII, were established that include proteins with truncated/ interrupted SET domains. Diverse mechanisms are involved in shaping the function and regulation of SET proteins. These include protein–protein interactions through both intra- and inter-molecular associations that are important in plant developmental processes, such as flowering time control and embryogenesis. Alternative splicing that can result in the generation of two to several different transcript isoforms is now known to be widespread. -

Automethylation of PRC2 Promotes H3K27 Methylation and Is Impaired in H3K27M Pediatric Glioma
Downloaded from genesdev.cshlp.org on October 5, 2021 - Published by Cold Spring Harbor Laboratory Press Automethylation of PRC2 promotes H3K27 methylation and is impaired in H3K27M pediatric glioma Chul-Hwan Lee,1,2,7 Jia-Ray Yu,1,2,7 Jeffrey Granat,1,2,7 Ricardo Saldaña-Meyer,1,2 Joshua Andrade,3 Gary LeRoy,1,2 Ying Jin,4 Peder Lund,5 James M. Stafford,1,2,6 Benjamin A. Garcia,5 Beatrix Ueberheide,3 and Danny Reinberg1,2 1Department of Biochemistry and Molecular Pharmacology, New York University School of Medicine, New York, New York 10016, USA; 2Howard Hughes Medical Institute, Chevy Chase, Maryland 20815, USA; 3Proteomics Laboratory, New York University School of Medicine, New York, New York 10016, USA; 4Shared Bioinformatics Core, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York 11724, USA; 5Department of Biochemistry and Molecular Biophysics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania 19104, USA The histone methyltransferase activity of PRC2 is central to the formation of H3K27me3-decorated facultative heterochromatin and gene silencing. In addition, PRC2 has been shown to automethylate its core subunits, EZH1/ EZH2 and SUZ12. Here, we identify the lysine residues at which EZH1/EZH2 are automethylated with EZH2-K510 and EZH2-K514 being the major such sites in vivo. Automethylated EZH2/PRC2 exhibits a higher level of histone methyltransferase activity and is required for attaining proper cellular levels of H3K27me3. While occurring inde- pendently of PRC2 recruitment to chromatin, automethylation promotes PRC2 accessibility to the histone H3 tail. Intriguingly, EZH2 automethylation is significantly reduced in diffuse intrinsic pontine glioma (DIPG) cells that carry a lysine-to-methionine substitution in histone H3 (H3K27M), but not in cells that carry either EZH2 or EED mutants that abrogate PRC2 allosteric activation, indicating that H3K27M impairs the intrinsic activity of PRC2. -

Human Catechol O-Methyltransferase Genetic Variation
Molecular Psychiatry (2004) 9, 151–160 & 2004 Nature Publishing Group All rights reserved 1359-4184/04 $25.00 www.nature.com/mp ORIGINAL RESEARCH ARTICLE Human catechol O-methyltransferase genetic variation: gene resequencing and functional characterization of variant allozymes AJ Shield1, BA Thomae1, BW Eckloff2, ED Wieben2 and RM Weinshilboum1 1Department of Molecular Pharmacology and Experimental Therapeutics, Mayo Medical School, Mayo Clinic, Mayo Foundation, Rochester, MN, USA; 2Department of Biochemistry and Molecular Biology, Mayo Medical School, Mayo Clinic, Mayo Foundation, Rochester, MN, USA Catechol O-methyltransferase (COMT) plays an important role in the metabolism of catecholamines, catecholestrogens and catechol drugs. A common COMT G472A genetic polymorphism (Val108/158Met) that was identified previously is associated with decreased levels of enzyme activity and has been implicated as a possible risk factor for neuropsychiatric disease. We set out to ‘resequence’ the human COMT gene using DNA samples from 60 African-American and 60 Caucasian-American subjects. A total of 23 single nucleotide polymorphisms (SNPs), including a novel nonsynonymous cSNP present only in DNA from African-American subjects, and one insertion/deletion were observed. The wild type (WT) and two variant allozymes, Thr52 and Met108, were transiently expressed in COS-1 and HEK293 cells. There was no significant change in level of COMT activity for the Thr52 variant allozyme, but there was a 40% decrease in the level of activity in cells transfected with the Met108 construct. Apparent Km values of the WT and variant allozymes for the two reaction cosubstrates differed slightly, but significantly, for 3,4-dihydroxybenzoic acid but not for S-adenosyl-L-methionine. -

Histone Methylases As Novel Drug Targets. Focus on EZH2 Inhibition. Catherine BAUGE1,2,#, Céline BAZILLE 1,2,3, Nicolas GIRARD1
Histone methylases as novel drug targets. Focus on EZH2 inhibition. Catherine BAUGE1,2,#, Céline BAZILLE1,2,3, Nicolas GIRARD1,2, Eva LHUISSIER1,2, Karim BOUMEDIENE1,2 1 Normandie Univ, France 2 UNICAEN, EA4652 MILPAT, Caen, France 3 Service d’Anatomie Pathologique, CHU, Caen, France # Correspondence and copy request: Catherine Baugé, [email protected], EA4652 MILPAT, UFR de médecine, Université de Caen Basse-Normandie, CS14032 Caen cedex 5, France; tel: +33 231068218; fax: +33 231068224 1 ABSTRACT Posttranslational modifications of histones (so-called epigenetic modifications) play a major role in transcriptional control and normal development, and are tightly regulated. Disruption of their control is a frequent event in disease. Particularly, the methylation of lysine 27 on histone H3 (H3K27), induced by the methylase Enhancer of Zeste homolog 2 (EZH2), emerges as a key control of gene expression, and a major regulator of cell physiology. The identification of driver mutations in EZH2 has already led to new prognostic and therapeutic advances, and new classes of potent and specific inhibitors for EZH2 show promising results in preclinical trials. This review examines roles of histone lysine methylases and demetylases in cells, and focuses on the recent knowledge and developments about EZH2. Key-terms: epigenetic, histone methylation, EZH2, cancerology, tumors, apoptosis, cell death, inhibitor, stem cells, H3K27 2 Histone modifications and histone code Epigenetic has been defined as inheritable changes in gene expression that occur without a change in DNA sequence. Key components of epigenetic processes are DNA methylation, histone modifications and variants, non-histone chromatin proteins, small interfering RNA (siRNA) and micro RNA (miRNA). -

Engaging Chromatin: PRC2 Structure Meets Function
www.nature.com/bjc REVIEW ARTICLE Engaging chromatin: PRC2 structure meets function Paul Chammas1, Ivano Mocavini1 and Luciano Di Croce1,2,3 Polycomb repressive complex 2 (PRC2) is a key epigenetic multiprotein complex involved in the regulation of gene expression in metazoans. PRC2 is formed by a tetrameric core that endows the complex with histone methyltransferase activity, allowing it to mono-, di- and tri-methylate histone H3 on lysine 27 (H3K27me1/2/3); H3K27me3 is a hallmark of facultative heterochromatin. The core complex of PRC2 is bound by several associated factors that are responsible for modulating its targeting specificity and enzymatic activity. Depletion and/or mutation of the subunits of this complex can result in severe developmental defects, or even lethality. Furthermore, mutations of these proteins in somatic cells can be drivers of tumorigenesis, by altering the transcriptional regulation of key tumour suppressors or oncogenes. In this review, we present the latest results from structural studies that have characterised PRC2 composition and function. We compare this information with data and literature for both gain-of function and loss-of-function missense mutations in cancers to provide an overview of the impact of these mutations on PRC2 activity. British Journal of Cancer (2020) 122:315–328; https://doi.org/10.1038/s41416-019-0615-2 BACKGROUND and embryonic ectoderm development (EED) (Table 1). These Transcriptional diversity is one of the hallmarks of cellular three proteins form the minimal core that confers histone identity. It is largely regulated at the level of chromatin, where methyltransferase (HMT) activity. A fourth factor, retinoblastoma- different protein complexes act as initiators, enhancers and/or binding protein (RBBP)4/7 (also known as RBAP48/46), has a repressors of transcription. -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

EZH2 in Normal Hematopoiesis and Hematological Malignancies
www.impactjournals.com/oncotarget/ Oncotarget, Vol. 7, No. 3 EZH2 in normal hematopoiesis and hematological malignancies Laurie Herviou2, Giacomo Cavalli2, Guillaume Cartron3,4, Bernard Klein1,2,3 and Jérôme Moreaux1,2,3 1 Department of Biological Hematology, CHU Montpellier, Montpellier, France 2 Institute of Human Genetics, CNRS UPR1142, Montpellier, France 3 University of Montpellier 1, UFR de Médecine, Montpellier, France 4 Department of Clinical Hematology, CHU Montpellier, Montpellier, France Correspondence to: Jérôme Moreaux, email: [email protected] Keywords: hematological malignancies, EZH2, Polycomb complex, therapeutic target Received: August 07, 2015 Accepted: October 14, 2015 Published: October 20, 2015 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Enhancer of zeste homolog 2 (EZH2), the catalytic subunit of the Polycomb repressive complex 2, inhibits gene expression through methylation on lysine 27 of histone H3. EZH2 regulates normal hematopoietic stem cell self-renewal and differentiation. EZH2 also controls normal B cell differentiation. EZH2 deregulation has been described in many cancer types including hematological malignancies. Specific small molecules have been recently developed to exploit the oncogenic addiction of tumor cells to EZH2. Their therapeutic potential is currently under evaluation. This review summarizes the roles of EZH2 in normal and pathologic hematological processes and recent advances in the development of EZH2 inhibitors for the personalized treatment of patients with hematological malignancies. PHYSIOLOGICAL FUNCTIONS OF EZH2 state through tri-methylation of lysine 27 on histone H3 (H3K27me3) [6]. -
Ordered Changes in Histone Modifications at the Core of The
Ordered changes in histone modifications at the core of the Arabidopsis circadian clock Jordi Malapeira1, Lucie Crhak Khaitova1, and Paloma Mas2 Molecular Genetics Department, Center for Research in Agricultural Genomics (CRAG), Consortium Consejo Superior de Investigaciones Científicas–Institut de Recerca i Tecnologia Agroalimentaries–Universitat Autònoma de Barcelona–Universitat de Barcelona, Campus Universitat Autònoma de Barcelona, 08193 Barcelona, Spain Edited* by Steve A. Kay, University of California at San Diego, La Jolla, CA, and approved November 15, 2012 (received for review October 1, 2012) Circadian clock function in Arabidopsis thaliana relies on a com- RELATED 3 (SDG2/ATXR3) was proposed to play a major role plex network of reciprocal regulations among oscillator components. in H3K4 trimethylation (H3K4me3) in Arabidopsis (16, 17). Loss Here, we demonstrate that chromatin remodeling is a prevalent of SDG2/ATXR3 function results in pleiotropic phenotypes, as well regulatory mechanism at the core of the clock. The peak-to-trough as a global decrease of H3K4me3 accumulation and altered ex- circadian oscillation is paralleled by the sequential accumulation of pression of a large number of genes. H3 acetylation (H3K56ac, K9ac), H3K4 trimethylation (H3K4me3), A precise regulation of gene expression is not only essential and H3K4me2. Inhibition of acetylation and H3K4me3 abolishes os- for plant responses to environmental stresses and developmental cillator gene expression, indicating that both marks are essential for transitions but also for proper function of the circadian clock. gene activation. Mechanistically, blocking H3K4me3 leads to in- The circadian clockwork allows plants to anticipate environ- creased clock-repressor binding, suggesting that H3K4me3 functions mental changes and adapt their activity to the most appropriate as a transition mark modulating the progression from activation to time of day (18). -

Epigenetic Regulation of Development by Histone Lysine Methylation
Heredity (2010) 105, 24–37 & 2010 Macmillan Publishers Limited All rights reserved 0018-067X/10 $32.00 www.nature.com/hdy REVIEW Epigenetic regulation of development by histone lysine methylation S Dambacher1, M Hahn1 and G Schotta Munich Center for Integrated Protein Science (CiPSM) and Adolf-Butenandt-Institute, Ludwig-Maximilians-University, Munich, Germany Epigenetic mechanisms contribute to the establishment and (HMTases) and specific binding factors for most methylated maintenance of cell-type-specific gene expression patterns. lysine positions has provided a novel insight into the mechanisms In this review, we focus on the functions of histone lysine of epigenetic gene regulation. In addition, analyses of HMTase methylation in the context of epigenetic gene regulation during knockout mice show that histone lysine methylation has developmental transitions. Over the past few years, analysis of important functions for normal development. In this study, we histone lysine methylation in active and repressive nuclear review mechanisms of gene activation and repression by histone compartments and, more recently, genome-wide profiling of lysine methylation and discuss them in the context of the histone lysine methylation in different cell types have revealed developmental roles of HMTases. correlations between particular modifications and the transcrip- Heredity (2010) 105, 24–37; doi:10.1038/hdy.2010.49; tional status of genes. Identification of histone methyltransferases published online 5 May 2010 Keywords: epigenetics; histone lysine methylation; heterochromatin; mouse development Introduction Activation and repression are facilitated by Development is accomplished by spatial and temporal histone lysine methylation regulation of gene expression patterns. The identity of Major methylation sites on histones H3 and H4 are each cell type is maintained and passed on to daughter located in the tail (H3K4, H3K9, H3K27, H3K36 and cells by mechanisms that do not alter the DNA sequence H4K20) and the nucleosome core region (H3K79). -

Generate Metabolic Map Poster
Authors: Pallavi Subhraveti Ron Caspi Peter Midford Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_003855395Cyc: Shewanella livingstonensis LMG 19866 Cellular Overview Connections between pathways are omitted for legibility. -
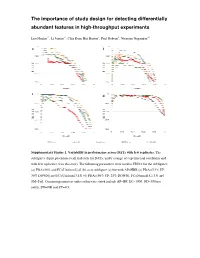
The Importance of Study Design Abundant Features in High
The importance of study design for detecting differentially abundant features in high -throughput experiments Luo Huaien 1* , Li Juntao 1*, C hia Kuan Hui Burton 1, Paul Robson 2, Niranjan Nagarajan 1# a b c d Supplementary Figure 1. Variability in performance across DATs with few repreplicates.licates. The subfigures depict precision-recall tradeoffs for DATs, under a range of experimexperimenentaltal conditions and with few replicates (2 in this case). The following parameters were used in EDDA for the sub figures: (a) PDA=30% and FC=Uniform[2,6] (b) as in subfigure (a) but with AP=HBR (c) PDA= [15% UP, 30% DOWN] and FC=Uniform[3,15] (d) PDA=[50% UP, 25% DOWN], FC=Normal[5,1.33] and SM=Full. Common parameters unless otherwise stated include AP=BP, EC=1000, ND=500 per entity, SM=NB and SV=0.5. a b c d Supplementary Figure 2 . Variability in performance across DATs with few data -points. Note that as in Supplementary Figure 1 , the performance of DATs varies widely merely by changing the number of replicates from 1 to 4 in sub figures (a)-(d) in a situation where the number data -points generated is limited (only 50 on average per entity). Results reported are based on data generated in EDDA with the following parameters (roughly mimicking a microbial RNA-seq application) : EC=1000, PDA=26%, FC=Uniform[3,7 ], AP=BP, SM=Full and SV=0.85. Supplementary Figure 3. Distribution of relative abundance in different empirically derived profiles. Note that both axes are plotted on a log-scale and that while BP does not have entities with relative abundance < 1e-5, the profile from Wu et al is more enriched for entities with relative abundance < 1e-5 than the HBR profile. -

Expanding the Structural Diversity of DNA Methyltransferase Inhibitors
pharmaceuticals Article Expanding the Structural Diversity of DNA Methyltransferase Inhibitors K. Eurídice Juárez-Mercado 1 , Fernando D. Prieto-Martínez 1 , Norberto Sánchez-Cruz 1 , Andrea Peña-Castillo 1, Diego Prada-Gracia 2 and José L. Medina-Franco 1,* 1 DIFACQUIM Research Group, Department of Pharmacy, School of Chemistry, National Autonomous University of Mexico, Avenida Universidad 3000, Mexico City 04510, Mexico; [email protected] (K.E.J.-M.); [email protected] (F.D.P.-M.); [email protected] (N.S.-C.); [email protected] (A.P.-C.) 2 Research Unit on Computational Biology and Drug Design, Children’s Hospital of Mexico Federico Gomez, Mexico City 06720, Mexico; [email protected] * Correspondence: [email protected] Abstract: Inhibitors of DNA methyltransferases (DNMTs) are attractive compounds for epigenetic drug discovery. They are also chemical tools to understand the biochemistry of epigenetic processes. Herein, we report five distinct inhibitors of DNMT1 characterized in enzymatic inhibition assays that did not show activity with DNMT3B. It was concluded that the dietary component theaflavin is an inhibitor of DNMT1. Two additional novel inhibitors of DNMT1 are the approved drugs glyburide and panobinostat. The DNMT1 enzymatic inhibitory activity of panobinostat, a known pan inhibitor of histone deacetylases, agrees with experimental reports of its ability to reduce DNMT1 activity in liver cancer cell lines. Molecular docking of the active compounds with DNMT1, and re-scoring with the recently developed extended connectivity interaction features approach, led to an excellent agreement between the experimental IC50 values and docking scores. Citation: Juárez-Mercado, K.E.; Keywords: dietary component; epigenetics; enzyme inhibition; focused library; epi-informatics; Prieto-Martínez, F.D.; Sánchez-Cruz, multitarget epigenetic agent; natural products; chemoinformatics N.; Peña-Castillo, A.; Prada-Gracia, D.; Medina-Franco, J.L.