Polyploidy and the Evolutionary History of Cotton Jonathan F
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Natural Materials for the Textile Industry Alain Stout
English by Alain Stout For the Textile Industry Natural Materials for the Textile Industry Alain Stout Compiled and created by: Alain Stout in 2015 Official E-Book: 10-3-3016 Website: www.TakodaBrand.com Social Media: @TakodaBrand Location: Rotterdam, Holland Sources: www.wikipedia.com www.sensiseeds.nl Translated by: Microsoft Translator via http://www.bing.com/translator Natural Materials for the Textile Industry Alain Stout Table of Contents For Word .............................................................................................................................. 5 Textile in General ................................................................................................................. 7 Manufacture ....................................................................................................................... 8 History ................................................................................................................................ 9 Raw materials .................................................................................................................... 9 Techniques ......................................................................................................................... 9 Applications ...................................................................................................................... 10 Textile trade in Netherlands and Belgium .................................................................... 11 Textile industry ................................................................................................................... -

Stable and Widespread Structural Heteroplasmy in Chloroplast Genomes Revealed by a New Long-Read Quantification Method
bioRxiv preprint doi: https://doi.org/10.1101/692798; this version posted July 11, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Classification: Biological Sciences, Evolution Title: Stable and widespread structural heteroplasmy in chloroplast genomes revealed by a new long-read quantification method Weiwen Wang a, Robert Lanfear a a Research School of Biology, Australian National University, Canberra, ACT, Australia, 2601 Corresponding Author: Weiwen Wang, [email protected] Robert Lanfear, [email protected], +61 2 6125 2536 Keywords: Single copy inversion, flip-flop recombination, chloroplast genome structural heteroplasmy 1 bioRxiv preprint doi: https://doi.org/10.1101/692798; this version posted July 11, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Abstract 2 The chloroplast genome usually has a quadripartite structure consisting of a large 3 single copy region and a small single copy region separated by two long inverted 4 repeats. It has been known for some time that a single cell may contain at least two 5 structural haplotypes of this structure, which differ in the relative orientation of the 6 single copy regions. However, the methods required to detect and measure the 7 abundance of the structural haplotypes are labour-intensive, and this phenomenon 8 remains understudied. -

Gossypium Barbadense: an Approach for in Situ Conservation in Cerrado, Brazil
Journal of Agricultural Science; Vol. 8, No. 8; 2016 ISSN 1916-9752 E-ISSN 1916-9760 Published by Canadian Center of Science and Education Gossypium barbadense: An Approach for in Situ Conservation in Cerrado, Brazil Andrezza Arantes Castro1, Lúcia Vieira Hoffmann2, Thiago Henrique Lima1, Aryanny Irene Domingos Oliveira1, Rafaela Ribeiro Brito1, Letícia de Maria Oliveira Mendes1, Caio César Oliveira Pereira1, Guilherme Malafaia1 & Ivandilson Pessoa Pinto de Menezes1 1 Genetic Molecular Laboratory, Instituto Federal Goiano, Urutaí, Goiás, Brazil 2 Embrapa Algodão, Campina Grande, Paraíba, Brazil Correspondence: Ivandilson Pessoa Pinto de Menezes, School Genetic Molecular Laboratory, Instituto Federal Goiano, Urutaí, Brazil. Tel: 55-64-9279-9708. E-mail: [email protected] Received: May 27, 2016 Accepted: June 16, 2016 Online Published: July 15, 2016 doi:10.5539/jas.v8n8p59 URL:http://dx.doi.org/10.5539/jas.v8n8p59 Abstract Abandonment of planting of Gossypium barbadense has endangered its existence. The objective was to determine the characteristicof the maintenance of Gossypium barbadense in the Central-West Region of Brazil, with the aim to foster the conservation of the species. Expeditions were conducted in 2014-2015 in Southeast Goiás, where cotton collection has not been reported before. Data from previous collections in Goiás, Mato Grosso, Mato Grosso do Sul and Distrito Federal available in Albrana database were considered this study. In the Central-West Region of Brazil, 466 accesses of G. barbadense were recorded, found most frequently in backyards (91.4%), but also spontaneous plants (7.5%), farm boundary (0.8%) and commercial farming (0.2%) have also been found. The main use indicated by VDU was as medicinal plant (0.66), therefore this is the main reason for in situ preservation. -

Complete Sequence of Kenaf (Hibiscus Cannabinus)
www.nature.com/scientificreports OPEN Complete sequence of kenaf (Hibiscus cannabinus) mitochondrial genome and comparative analysis Received: 2 November 2017 Accepted: 27 July 2018 with the mitochondrial genomes of Published: xx xx xxxx other plants Xiaofang Liao1,2,3, Yanhong Zhao3, Xiangjun Kong2, Aziz Khan2, Bujin Zhou 2, Dongmei Liu4, Muhammad Haneef Kashif2, Peng Chen2, Hong Wang5 & Ruiyang Zhou2 Plant mitochondrial (mt) genomes are species specifc due to the vast of foreign DNA migration and frequent recombination of repeated sequences. Sequencing of the mt genome of kenaf (Hibiscus cannabinus) is essential for elucidating its evolutionary characteristics. In the present study, single- molecule real-time sequencing technology (SMRT) was used to sequence the complete mt genome of kenaf. Results showed that the complete kenaf mt genome was 569,915 bp long and consisted of 62 genes, including 36 protein-coding, 3 rRNA and 23 tRNA genes. Twenty-fve introns were found among nine of the 36 protein-coding genes, and fve introns were trans-spliced. A comparative analysis with other plant mt genomes showed that four syntenic gene clusters were conserved in all plant mtDNAs. Fifteen chloroplast-derived fragments were strongly associated with mt genes, including the intact sequences of the chloroplast genes psaA, ndhB and rps7. According to the plant mt genome evolution analysis, some ribosomal protein genes and succinate dehydrogenase genes were frequently lost during the evolution of angiosperms. Our data suggest that the kenaf mt genome retained evolutionarily conserved characteristics. Overall, the complete sequencing of the kenaf mt genome provides additional information and enhances our better understanding of mt genomic evolution across angiosperms. -

Characterization of Some Common Members of the Family Malvaceae S.S
Indian Journal of Plant Sciences ISSN: 2319–3824(Online) An Open Access, Online International Journal Available at http://www.cibtech.org/jps.htm 2014 Vol. 3 (3) July-September, pp.79-86/Naskar and Mandal Research Article CHARACTERIZATION OF SOME COMMON MEMBERS OF THE FAMILY MALVACEAE S.S. ON THE BASIS OF MORPHOLOGY OF SELECTIVE ATTRIBUTES: EPICALYX, STAMINAL TUBE, STIGMATIC HEAD AND TRICHOME *Saikat Naskar and Rabindranath Mandal Department of Botany, Barasat Govt. College, Barasat, Kolkata- 700124, West Bengal, India *Author for Correspondence: [email protected] ABSTRACT Epicalyx, staminal tube, stigma and trichome morphological characters have been used to characterize some common members of Malvaceae s.s. These characters have been analyzed following a recent molecular phylogenetic classification of Malvaceae s.s. Stigmatic character is effective for segregation of the tribe Gossypieae from other tribes. But precise distinction of other two studied tribes, viz. Hibisceae and Malveae on the basis of this character proved to be insufficient. Absence of epicalyx in Malachra has indicated an independent evolutionary event within Hibisceae. Distinct H-shaped trichome of Malvastrum has pointed out its isolated position within Malveae. Staminal tube morphological similarities of Abutilon and Sida have suggested their closeness. A key to the genera has been provided for identification purpose. Keywords: Malvaceae s.s., Epicalyx, Staminal Tube, Stigma, Trichome INTRODUCTION Epicalyx and monadelphous stamens are considered as key characters of the family Malvaceae s.s. Epicalyx was recognized as an important character for taxonomic value by several authors (Fryxell, 1988; Esteves, 2000) since its presence or absence was employed to determine phylogenetic interpretation within the tribes of Malvaceae s.s. -
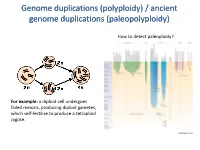
Polyploidy) / Ancient Genome Duplications (Paleopolyploidy
Genome duplications (polyploidy) / ancient genome duplications (paleopolyploidy) How to detect paleoploidy? For example: a diploid cell undergoes failed meiosis, producing diploid gametes, which self-fertilize to produce a tetraploid zygote. Timing of duplication by trees (phylogenetic timing) Phylogenetic timing of duplicates b Paramecium genome duplications Comparison of two scaffolds originating from a common ancestor at the recent WGD Saccharomyces cerevisiae Just before genome duplication Just after genome duplication More time after genome duplication Unaligned view (removing gaps just like in cerev has occurred) Saccharomyces cerevisiae Problem reciprocal gene loss (extreme case); how to solve? Problem reciprocal gene loss (extreme case); how to solve? Just before genome duplication Outgroup! Just after genome duplication Outgroup Just after genome duplication Outgroup More time after genome duplication Outgroup Problem (extreme case); how to solve? Outgroup Outgroup Outgroup Outgroup Outgroup Using other genomes Wong et al. 2002 PNAS Centromeres Vertebrate genome duplication Nature. 2011 Apr 10. [Epub ahead of print] Ancestral polyploidy in seed plants and angiosperms. Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, Soltis DE, Clifton SW, Schlarbaum SE, Schuster SC, Ma H, Leebens-Mack J, Depamphilis CW. Flowering plants Flowering MOSS Vertebrates Teleosts S. serevisiae and close relatives Paramecium Reconstructed map of genome duplications allows unprecedented mapping -

Polyploidy and the Evolutionary History of Cotton
POLYPLOIDY AND THE EVOLUTIONARY HISTORY OF COTTON Jonathan F. Wendel1 and Richard C. Cronn2 1Department of Botany, Iowa State University, Ames, Iowa 50011, USA 2Pacific Northwest Research Station, USDA Forest Service, 3200 SW Jefferson Way, Corvallis, Oregon 97331, USA I. Introduction II. Taxonomic, Cytogenetic, and Phylogenetic Framework A. Origin and Diversification of the Gossypieae, the Cotton Tribe B. Emergence and Diversification of the Genus Gossypium C. Chromosomal Evolution and the Origin of the Polyploids D. Phylogenetic Relationships and the Temporal Scale of Divergence III. Speciation Mechanisms A. A Fondness for Trans-oceanic Voyages B. A Propensity for Interspecific Gene Exchange IV. Origin of the Allopolyploids A. Time of Formation B. Parentage of the Allopolyploids V. Polyploid Evolution A. Repeated Cycles of Genome Duplication B. Chromosomal Stabilization C. Increased Recombination in Polyploid Gossypium D. A Diverse Array of Genic and Genomic Interactions E. Differential Evolution of Cohabiting Genomes VI. Ecological Consequences of Polyploidization VII. Polyploidy and Fiber VIII. Concluding Remarks References The cotton genus (Gossypium ) includes approximately 50 species distributed in arid to semi-arid regions of the tropic and subtropics. Included are four species that have independently been domesticated for their fiber, two each in Africa–Asia and the Americas. Gossypium species exhibit extraordinary morphological variation, ranging from herbaceous perennials to small trees with a diverse array of reproductive and vegetative -

The Diversity of Plant Sex Chromosomes Highlighted Through Advances in Genome Sequencing
G C A T T A C G G C A T genes Review The Diversity of Plant Sex Chromosomes Highlighted through Advances in Genome Sequencing Sarah Carey 1,2 , Qingyi Yu 3,* and Alex Harkess 1,2,* 1 Department of Crop, Soil, and Environmental Sciences, Auburn University, Auburn, AL 36849, USA; [email protected] 2 HudsonAlpha Institute for Biotechnology, Huntsville, AL 35806, USA 3 Texas A&M AgriLife Research, Texas A&M University System, Dallas, TX 75252, USA * Correspondence: [email protected] (Q.Y.); [email protected] (A.H.) Abstract: For centuries, scientists have been intrigued by the origin of dioecy in plants, characterizing sex-specific development, uncovering cytological differences between the sexes, and developing theoretical models. Through the invention and continued improvements in genomic technologies, we have truly begun to unlock the genetic basis of dioecy in many species. Here we broadly review the advances in research on dioecy and sex chromosomes. We start by first discussing the early works that built the foundation for current studies and the advances in genome sequencing that have facilitated more-recent findings. We next discuss the analyses of sex chromosomes and sex-determination genes uncovered by genome sequencing. We synthesize these results to find some patterns are emerging, such as the role of duplications, the involvement of hormones in sex-determination, and support for the two-locus model for the origin of dioecy. Though across systems, there are also many novel insights into how sex chromosomes evolve, including different sex-determining genes and routes to suppressed recombination. We propose the future of research in plant sex chromosomes should involve interdisciplinary approaches, combining cutting-edge technologies with the classics Citation: Carey, S.; Yu, Q.; to unravel the patterns that can be found across the hundreds of independent origins. -

Genetic Variability Studies in Gossypium Barbadense L
Electronic Journal of Plant Breeding, 1(4): 961-965 (July 2010) Research Article Genetic variability studies in Gossypium barbadense L. genotypes for seed cotton yield and its yield components K. P. M. Dhamayanathi , S. Manickam and K. Rathinavel Abstract A study was carried out during kharif 2006-07 with twenty five Gossypium barbadense L genotypes to obtain information on genetic variability, heritability and genetic advance for seed cotton yield and its yield attributes. Significant differences were observed for characters among genotypes. High genetic differences were recorded for nodes/plant, sympodia, bolls as well as fruiting points per plant, seed cotton yield, lint index indicating ample scope for genetic improvement of these characters through selection. Results also revealed high heritability coupled with high genetic advance for yield and most of the yield components as well as fibre quality traits. Sympodia/plant, fruiting point /plant, number of nodes/plant, number of bolls per plant, and lint index were positively correlated with seed cotton yield per plant and appeared to be interrelated with each other. It is suggested that these characters could be considered as selection criteria in improving the seed cotton yield of G. barbadense , L genotypes. Key words : Gossypium barbadense , genetic variability, heritability, genetic advance, lint index, selection criteria Introduction Seed cotton yield is a complex trait governed by Cotton is the most widely used vegetable fibre and several yield contributing characters such as plant also the most important raw material for the textile height, number of monopodia, number of industry, grown in tropical and subtropical regions sympodia, number of bolls, number of fruiting in more than 80 countries all over the world. -

Long-Reads Reveal That the Chloroplast Genome Exists in Two Distinct Versions in Most Plants
GBE Long-Reads Reveal That the Chloroplast Genome Exists in Two Distinct Versions in Most Plants Weiwen Wang* and Robert Lanfear* Division of Ecology and Evolution, Research School of Biology, Australian National University, Acton, Australian Capital Territory, Australia *Corresponding authors: E-mails: [email protected]; [email protected]. Accepted: November 15, 2019 Downloaded from https://academic.oup.com/gbe/article/11/12/3372/5637229 by guest on 02 October 2021 Data deposition: The Herrania umbratica and Siraitia grosvenorii chloroplast genomes in this project have been deposited at NCBI under the accession MN163033 and MK279915. Abstract The chloroplast genome usually has a quadripartite structure consisting of a large single copy region and a small single copy region separated by two long inverted repeats. It has been known for some time that a single cell may contain at least two structural haplotypes of this structure, which differ in the relative orientation of the single copy regions. However, the methods required to detect and measure the abundance of the structural haplotypes are labor-intensive, and this phenomenon remains understudied. Here, we develop a new method, Cp-hap, to detect all possible structural haplotypes of chloroplast genomes of quadripartite structure using long-read sequencing data. We use this method to conduct a systematic analysis and quantification of chloroplast structural haplotypes in 61 land plant species across 19 orders of Angiosperms, Gymnosperms, and Pteridophytes. Our results show that there are two chloroplast structural haplotypes which occur with equal frequency in most land plant individuals. Nevertheless, species whose chloroplast genomes lack inverted repeats or have short inverted repeats have just a single structural haplotype. -

Methods to Enable the Coexistence of Diverse Cotton Production Systems
AGRICULTURAL BIOTECHNOLOGY IN CALIFORNIA SERIES PUBLICATION 8191 Methods to Enable the Coexistence of Diverse Cotton Production Systems ROBERT B. HUTMACHER, Extension Agronomist, University of California Shafter Research and Extension Center and University of California, Davis, Department of Plant Science; RON N. VARGAS, County Director and Farm Advisor, University of California Cooperative UNIVERSITY OF Extension, Madera and Merced Counties; STEVEN D. WRIGHT, Farm Advisor, University of CALIFORNIA California Cooperative Extension, Tulare and Kings Counties Division of Agriculture Upland cotton (Gossypium hirsutum) and Pima cotton (G. barbadense) are the two and Natural Resources types of cotton produced commercially in California. In acreage as well as crop http://anrcatalog.ucdavis.edu value, over the past 5 years cotton has typically ranked in the top three in agronomic field crops grown in California. During that period, plantings of upland cotton in California have ranged from about 400,000 to over 650,000 acres (160,000 to 260,000 ha), while Pima plantings have ranged from about 140,000 to over 250,000 acres (56,000 to 101,000 ha). Does cross-pollination occur in cotton? Both upland and Pima cotton are variously referred to as “largely self-pollinated” or “partially cross-pollinated.” These descriptions acknowledge that these types of cotton are mostly self-pollinated but some cross-pollination can occur, albeit at relatively low incidence rates, through activity of pollinating insects or by wind dispersion. The pol- len of both wild and cultivated Gossypium species is large in size and among the heaviest among angiosperms, the group of plants that produces flowers, fruit, and seeds. -

Variation Than Mainland Populations?
Heredity 78 (1997) 311—327 Received 30Apr11 1996 Do island populations have less genetic variation than mainland populations? R. FRANKHAM* Key Centre for Biodiversity and Bioresources, Macquarie University, Sydney, NSW2109, Australia Islandpopulations are much more prone to extinction than mainland populations. The reasons for this remain controversial. If inbreeding and loss of genetic variation are involved, then genetic variation must be lower on average in island than mainland populations. Published data on levels of genetic variation for allozymes, nuclear DNA markers, mitochondrial DNA, inversions and quantitative characters in island and mainland populations were analysed. A large and highly significant majority of island populations have less allozyme genetic variation than their mainland counterparts (165 of 202 comparisons), the average reduction being 29 per cent. The magnitude of differences was related to dispersal ability. There were related differ- ences for all the other measures. Island endemic species showed lower genetic variation than related mainland species in 34 of 38 cases. The proportionate reduction in genetic variation was significantly greater in island endemic than in nonendemic island populations in mammals and birds, but not in insects. Genetic factors cannot be discounted as a cause of higher extinction rates of island than mainland populations. Keywords:allozymes,conservation, endemic species, extinction, genetic variation, islands. of endemic plant species as threatened on 15 islands. Introduction Human activities have been the major cause of Islandpopulations have a much higher risk of species extinctions on islands in the past 50000 years extinction than mainland populations (Diamond, (Olson, 1989) through over-exploitation, habitat loss 1984; Vitousek, 1988; Flesness, 1989; Case et al., and introduced species.