Deep Learning in Science Pierre Baldi Frontmatter More Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Principled Design of Large-Scale Recursive Neural Network Architectures–DAG-Rnns and the Protein Structure Prediction Problem
Journal of Machine Learning Research 4 (2003) 575-602 Submitted 2/02; Revised 4/03; Published 9/03 The Principled Design of Large-Scale Recursive Neural Network Architectures–DAG-RNNs and the Protein Structure Prediction Problem Pierre Baldi [email protected] Gianluca Pollastri [email protected] School of Information and Computer Science Institute for Genomics and Bioinformatics University of California, Irvine Irvine, CA 92697-3425, USA Editor: Michael I. Jordan Abstract We describe a general methodology for the design of large-scale recursive neural network architec- tures (DAG-RNNs) which comprises three fundamental steps: (1) representation of a given domain using suitable directed acyclic graphs (DAGs) to connect visible and hidden node variables; (2) parameterization of the relationship between each variable and its parent variables by feedforward neural networks; and (3) application of weight-sharing within appropriate subsets of DAG connec- tions to capture stationarity and control model complexity. Here we use these principles to derive several specific classes of DAG-RNN architectures based on lattices, trees, and other structured graphs. These architectures can process a wide range of data structures with variable sizes and dimensions. While the overall resulting models remain probabilistic, the internal deterministic dy- namics allows efficient propagation of information, as well as training by gradient descent, in order to tackle large-scale problems. These methods are used here to derive state-of-the-art predictors for protein structural features such as secondary structure (1D) and both fine- and coarse-grained contact maps (2D). Extensions, relationships to graphical models, and implications for the design of neural architectures are briefly discussed. -

Methodology for Predicting Semantic Annotations of Protein Sequences by Feature Extraction Derived of Statistical Contact Potentials and Continuous Wavelet Transform
Universidad Nacional de Colombia Sede Manizales Master’s Thesis Methodology for predicting semantic annotations of protein sequences by feature extraction derived of statistical contact potentials and continuous wavelet transform Author: Supervisor: Gustavo Alonso Arango Dr. Cesar German Argoty Castellanos Dominguez A thesis submitted in fulfillment of the requirements for the degree of Master’s on Engineering - Industrial Automation in the Department of Electronic, Electric Engineering and Computation Signal Processing and Recognition Group June 2014 Universidad Nacional de Colombia Sede Manizales Tesis de Maestr´ıa Metodolog´ıapara predecir la anotaci´on sem´antica de prote´ınaspor medio de extracci´on de caracter´ısticas derivadas de potenciales de contacto y transformada wavelet continua Autor: Tutor: Gustavo Alonso Arango Dr. Cesar German Argoty Castellanos Dominguez Tesis presentada en cumplimiento a los requerimientos necesarios para obtener el grado de Maestr´ıaen Ingenier´ıaen Automatizaci´onIndustrial en el Departamento de Ingenier´ıaEl´ectrica,Electr´onicay Computaci´on Grupo de Procesamiento Digital de Senales Enero 2014 UNIVERSIDAD NACIONAL DE COLOMBIA Abstract Faculty of Engineering and Architecture Department of Electronic, Electric Engineering and Computation Master’s on Engineering - Industrial Automation Methodology for predicting semantic annotations of protein sequences by feature extraction derived of statistical contact potentials and continuous wavelet transform by Gustavo Alonso Arango Argoty In this thesis, a method to predict semantic annotations of the proteins from its primary structure is proposed. The main contribution of this thesis lies in the implementation of a novel protein feature representation, which makes use of the pairwise statistical contact potentials describing the protein interactions and geometry at the atomic level. -

Deep Learning in Chemoinformatics Using Tensor Flow
UC Irvine UC Irvine Electronic Theses and Dissertations Title Deep Learning in Chemoinformatics using Tensor Flow Permalink https://escholarship.org/uc/item/963505w5 Author Jain, Akshay Publication Date 2017 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA, IRVINE Deep Learning in Chemoinformatics using Tensor Flow THESIS submitted in partial satisfaction of the requirements for the degree of MASTER OF SCIENCE in Computer Science by Akshay Jain Thesis Committee: Professor Pierre Baldi, Chair Professor Cristina Videira Lopes Professor Eric Mjolsness 2017 c 2017 Akshay Jain DEDICATION To my family and friends. ii TABLE OF CONTENTS Page LIST OF FIGURES v LIST OF TABLES vi ACKNOWLEDGMENTS vii ABSTRACT OF THE THESIS viii 1 Introduction 1 1.1 QSAR Prediction Methods . .2 1.2 Deep Learning . .4 2 Artificial Neural Networks(ANN) 5 2.1 Artificial Neuron . .5 2.2 Activation Function . .7 2.3 Loss function . .8 2.4 Optimization . .8 3 Deep Recursive Architectures 10 3.1 Recurrent Neural Networks (RNN) . 10 3.2 Recursive Neural Networks . 11 3.3 Directed Acyclic Graph Recursive Neural Networks (DAG-RNN) . 11 4 UG-RNN for small molecules 14 4.1 DAG Generation . 16 4.2 Local Information Vector . 16 4.3 Contextual Vectors . 17 4.4 Activity Prediction . 17 4.5 UG-RNN With Contracted Rings (UG-RNN-CR) . 18 4.6 Example: UG-RNN Model of Propionic Acid . 20 5 Implementation 24 6 Data & Results 26 6.1 Aqueous Solubility Prediction . 26 6.2 Melting Point Prediction . 28 iii 7 Conclusions 30 Bibliography 32 A Source Code 37 A.1 UGRNN . -

Course Outline
Department of Computer Science and Software Engineering COMP 6811 Bioinformatics Algorithms (Reading Course) Fall 2019 Section AA Instructor: Gregory Butler Curriculum Description COMP 6811 Bioinformatics Algorithms (4 credits) The principal objectives of the course are to cover the major algorithms used in bioinformatics; sequence alignment, multiple sequence alignment, phylogeny; classi- fying patterns in sequences; secondary structure prediction; 3D structure prediction; analysis of gene expression data. This includes dynamic programming, machine learning, simulated annealing, and clustering algorithms. Algorithmic principles will be emphasized. A project is required. Outline of Topics The course will focus on algorithms for protein sequence analysis. It will not cover genome assembly, genome mapping, or gene recognition. • Background in Biology and Genomics • Sequence Alignment: Pairwise and Multiple • Representation of Protein Amino Acid Composition • Profile Hidden Markov Models • Specificity Determining Sites • Curation, Annotation, and Ontologies • Machine Learning: Secondary Structure, Signals, Subcellular Location • Protein Families, Phylogenomics, and Orthologous Groups • Profile-Based Alignments • Algorithms Based on k-mers 1 Texts | in Library D. Higgins and W. Taylor (editors). Bioinformatics: Sequence, Structure and Databanks, Oxford University Press, 2000. A. D. Baxevanis and B. F. F. Ouelette. Bioinformatics: A Practical Guide to the Analysis of Genes and Proteins, Wiley, 1998. Richard Durbin, Sean R. Eddy, Anders Krogh, -

Bringing Folding Pathways Into Strand Pairing Prediction
Bringing folding pathways into strand pairing prediction Jieun Jeong1,2, Piotr Berman1, and Teresa Przytycka2 1 Computer Science and Engineering Department The Pennsylvania State University University Park, PA 16802 USA 2 National Center for Biotechnology Information US National Library of Medicine, National Institutes of Health Bethesda, MD 20894 email: [email protected], [email protected], [email protected] Abstract. The topology of β-sheets is defined by the pattern of hydrogen- bonded strand pairing. Therefore, predicting hydrogen bonded strand partners is a fundamental step towards predicting β-sheet topology. In this work we report a new strand pairing algorithm. Our algorithm at- tempts to mimic elements of the folding process. Namely, in addition to ensuring that the predicted hydrogen bonded strand pairs satisfy basic global consistency constraints, it takes into account hypothetical folding pathways. Consistently with this view, introducing hydrogen bonds be- tween a pair of strands changes the probabilities of forming other strand pairs. We demonstrate that this approach provides an improvement over previously proposed algorithms. 1 Introduction The prediction of protein structure from protein sequence is a long-held goal that would provide invaluable information regarding the function of individ- ual proteins and the evolution of protein families. The increasing amount of sequence and structure data, made it possible to decouple the structure predic- tion problem from the problem of modeling of protein folding process. Indeed, a significant progress has been achieved by bioinformatics approaches such as homology modeling, threading, and assembly from fragments [16]. At the same time, the fundamental problem of how actually a protein acquires its final folded state remains a subject of controversy. -
ACDL-2020-Programme-Ver10.0
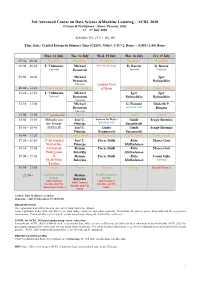
3rd Advanced Course on Data Science &Machine Learning – ACDL 2020 Certosa di Pontignano - Siena, Tuscany, Italy 13 – 17 July 2020 Schedule Ver. 11.0 – July 9th Time Zone: Central European Summer Time (CEST), Offset: UTC+2, Rome – (GMT+2:00) Rome Mon, 13 July Tue, 14 July Wed, 15 July Thu, 16 July Fri, 17 July 07:30 – 09:00 Breakfast Breakfast Breakfast Breakfast Breakfast 09:00 – 09:50 T. Viehmann Michael 8:30 Social Tour D. Bacciu D. Bacciu Tutorial Bronstein Tutorial Tutorial 09:50 – 10:40 Michael Igor Bronstein Babuschkin Tutorial Guided Visit 10:40 – 11:20 Coffee break Coffee break of Siena Coffee break Coffee break 11:20 – 12:10 T. Viehmann Michael Igor Igor Tutorial Bronstein Babuschkin Babuschkin Tutorial 12:10 – 13:00 Michael G. Fiameni Diederik P. Bronstein Industrial Talk Kingma Tutorial 13:00 – 15:00 Lunch (13:00-14:00) Lunch Lunch Lunch Lunch 15:00 – 15:50 Mihaela van José C. Lorenzo De Mattei Guido Sergiy Butenko der Schaar Principe Industrial Talk Sanguinetti 15:50 – 16:40 (14:00-16:40) José C. Guido Guido Sergiy Butenko Principe Sanguinetti Sanguinetti 16:40 – 17:20 Coffee break Coffee break Coffee break Coffee break Coffee break 17:20 – 18:10 17:20 Guided José C. Pierre Baldi Risto Marco Gori Visit of the Principe Miikkulainen 18:10 – 19:00 Certosa di Roman Pierre Baldi Risto Marco Gori Pontignano Belavkin Miikkulainen 19:00 – 19:50 & Roman Pierre Baldi Risto Varun Ojha 18:20 Wine Belavkin Miikkulainen Tutorial Tasting 19:50 – 21:50 Dinner Dinner Dinner Dinner Social Dinner 21:50 – Oral Presentation Roman Oral Presentation Session Belavkin Session with Cantucci with Cantucci with Cantucci biscuits and Vin biscuits and Vin biscuits and Vin Santo (sweet wine) Santo Santo Arrival: July 12 (Dinner at 20:30) Departure: July 18 (Breakfast 07:30-09:00) REGISTRATION The registration desk will be located close to the Main Conference Room. -

I S C B N E W S L E T T
ISCB NEWSLETTER FOCUS ISSUE {contents} President’s Letter 2 Member Involvement Encouraged Register for ISMB 2002 3 Registration and Tutorial Update Host ISMB 2004 or 2005 3 David Baker 4 2002 Overton Prize Recipient Overton Endowment 4 ISMB 2002 Committees 4 ISMB 2002 Opportunities 5 Sponsor and Exhibitor Benefits Best Paper Award by SGI 5 ISMB 2002 SIGs 6 New Program for 2002 ISMB Goes Down Under 7 Planning Underway for 2003 Hot Jobs! Top Companies! 8 ISMB 2002 Job Fair ISCB Board Nominations 8 Bioinformatics Pioneers 9 ISMB 2002 Keynote Speakers Invited Editorial 10 Anna Tramontano: Bioinformatics in Europe Software Recommendations11 ISCB Software Statement volume 5. issue 2. summer 2002 Community Development 12 ISCB’s Regional Affiliates Program ISCB Staff Introduction 12 Fellowship Recipients 13 Awardees at RECOMB 2002 Events and Opportunities 14 Bioinformatics events world wide INTERNATIONAL SOCIETY FOR COMPUTATIONAL BIOLOGY A NOTE FROM ISCB PRESIDENT This newsletter is packed with information on development and dissemination of bioinfor- the ISMB2002 conference. With over 200 matics. Issues arise from recommendations paper submissions and over 500 poster submis- made by the Society’s committees, Board of sions, the conference promises to be a scientific Directors, and membership at large. Important feast. On behalf of the ISCB’s Directors, staff, issues are defined as motions and are discussed EXECUTIVE COMMITTEE and membership, I would like to thank the by the Board of Directors on a bi-monthly Philip E. Bourne, Ph.D., President organizing committee, local organizing com- teleconference. Motions that pass are enacted Michael Gribskov, Ph.D., mittee, and program committee for their hard by the Executive Committee which also serves Vice President work preparing for the conference. -

Pierre Baldi
Pierre Baldi School of Information and Computer Sciences Department of Computer Science Department of Biological Chemistry Institute for Genomics and Bioinformatics California Institute for Telecommunications and Information Technology University of California, Irvine Irvine, CA 92697-3435 (949) 824-5809 (949) 824-9813 FAX Assistant: Katarina Fletcher ([email protected]) [email protected] www.ics.uci.edu/~pfbaldi www.igb.uci.edu Professional Experience • October 2006 to present: Chancellor’s Professor • September 2006 to present: Associate-Director, Center for Machine Learning and Intelligent Systems. • January 2001 to present: Director Institute for Genomics and Bioinformatics. • June 2001 to present: Professor Department of Information and Computer Science, University of California, Irvine. [Joint appointment in the Department of Biological Chemistry, College of Medicine and the Department of Developmental and Cell Biology, School of Biological Sciences]. • January 2002 to December 2005: Application Layer Leader (Digitally Enabled Medicine) for the California Institute for Telecommunications and Information Technology [Calit2 2]. • July 1999 to May 2001: Associate Professor, Department of Information and Computer Science, University of California, Irvine. [Joint appointment in the Department of Biological Chemistry, College of Medicine and the Department of Developmental and Cell Biology, School of Biological Sciences]. • 1991 to June 1999: Chairman and CEO, Net-ID, Inc. • January 1999: Visiting Professor, Department of Computer Science, University of Florence. • 1995 to 1996: Member of the Professional Staff, Division of Biology, California Institute of Technology. • 1988 to 1995: Member of the Technical Staff in the Nonlinear Science and Information Processing Group at the Jet Propulsion Laboratory, and Visiting Research Associate, Division of Biology, California Institute of Technology. -
Top 100 AI Leaders in Drug Discovery and Advanced Healthcare Introduction
Top 100 AI Leaders in Drug Discovery and Advanced Healthcare www.dka.global Introduction Over the last several years, the pharmaceutical and healthcare organizations have developed a strong interest toward applying artificial intelligence (AI) in various areas, ranging from medical image analysis and elaboration of electronic health records (EHRs) to more basic research like building disease ontologies, preclinical drug discovery, and clinical trials. The demand for the ML/AI technologies, as well as for ML/AI talent, is growing in pharmaceutical and healthcare industries and driving the formation of a new interdisciplinary industry (‘data-driven healthcare’). Consequently, there is a growing number of AI-driven startups and emerging companies offering technology solutions for drug discovery and healthcare. Another important source of advanced expertise in AI for drug discovery and healthcare comes from top technology corporations (Google, Microsoft, Tencent, etc), which are increasingly focusing on applying their technological resources for tackling health-related challenges, or providing technology platforms on rent bases for conducting research analytics by life science professionals. Some of the leading pharmaceutical giants, like GSK, AstraZeneca, Pfizer and Novartis, are already making steps towards aligning their internal research workflows and development strategies to start embracing AI-driven digital transformation at scale. However, the pharmaceutical industry at large is still lagging behind in adopting AI, compared to more traditional consumer industries -- finance, retail etc. The above three main forces are driving the growth in the AI implementation in pharmaceutical and advanced healthcare research, but the overall success depends strongly on the availability of highly skilled interdisciplinary leaders, able to innovate, organize and guide in this direction. -

Predicting Protein Folding Pathways Using Ensemble Modeling and Sequence Information
Predicting Protein Folding Pathways Using Ensemble Modeling and Sequence Information by David Becerra McGill University School of Computer Science Montr´eal, Qu´ebec, Canada A thesis submitted to McGill University in partial fulfillment of the requirements of the degree of Doctor of Philosophy c David Becerra 2017 Dedicated to my GRANDMOM. Thanks for everything. You are such a strong woman i Acknowledgements My doctoral program has been an amazing adventure with ups and downs. The most valuable fact is that I am a different person with respect to the one who started the program five years ago. I am not better or worst, I am just different. I would like to express my gratitude to all the people who helped me during my time at McGill. This thesis contains only my name as author, but it should have the name of all the people who played an important role in the completion of this thesis (and they are a lot of people). I am certain I would have not reached the end of my thesis without the help, advises and support of all people around me. First at all, I want to thank my country Colombia. Being abroad, I learnt to value my culture, my roots and my south-american positiveness, happiness and resourcefulness. I am really proud of being Colombian and of showing to the world a bit of what our wonderful land has given to us. I would also like to thank all Colombians, because with your taxes (via Colciencia’s funding) I was able to complete this dream. -

PSB 2009 Attendees (As of January 9, 2009) Mario Albrecht Max Planck
PSB 2009 Attendees (as of January 9, 2009) Mario Albrecht Steven Brenner Max Planck Institute for Informatics University of California, Berkeley Hesham Ali Andrzej Brodzik University of Nebraska at Omaha The MITRE Corporation Gil Alterovitz Lukas Burger Harvard/MIT Institute of Bioinformatics, University of Basel Gregory Arnold Amgen Gregory Burrows OHSU Adam Asare UCSF/ITN William Bush Vanderbilt University Pierre Baldi University of California Andrea Califano Columbia University Lars Barquist UC Berkeley J. Gregory Caporaso Univeristy of Colorado Denver James Bassingthwaighte Univeristy of Washington Vincent Carey Harvard Medical School Tanya Berger-Wolf University of Illinois at Chicago Yuhui Cheng University of California, San Diego Ghislain Bidaut INSERM U891 Institut Paoli-Calmette Raymond Cheong Johns Hopkins University Marco Blanchette Stowers Institute for Medical Research Annie Chiang Stanford University Ben Blencowe Gilsoo Cho Anthony Bonner Yonsei University University of Toronto Sung Bum Cho Ingrid Borecki Seoul National University Bioinformatics Washington University Kevin Cohen John Bouck University of Colorado Denver Ceres, Inc. Dilek Colak Philip Bourne King Faisal Specialist Hospital and University of California, San Diego Research Centre PSB 2009 Attendees (as of January 9, 2009) James Costello Laura Elnitski Indiana University NHGRI Anneleen Daemen Drew Endy Dept. Electrical Engineering, KULeuven Stanford University Denise Daley Eric Fahrenthold University of British Columbia Yiping Fan Paul Davis St Jude Children's -

Digitally Enabled Genomic Medicine
digitally enabled genomic medicine fusing revolutions in genome-enabled biomedicine and the use of the internet and information technology in society projects and people at the university of california, san diego and cal-( it) 2 Cal-(IT)2’s mission is simple: Extend the reach of the Internet throughout the physical world. Cal-(IT)2 teams University of California, San Diego (UCSD) and University of California, Irvine (UCI), faculty, students, and research professionals with leading California telecommunications, computer, software, and applications companies to conduct research on the scientific and technological components needed to bring this new Internet into being. Institute applications researchers are conduct- ing their studies in “living laboratories” to investigate how this future Internet will accelerate advances in environmental sci- ence, civil infrastructure, intelligent transportation and telematics, genomic medicine, the new media arts, and educational practices. Cal-(IT)2’s research program is organized conceptually into vertically interlocking and collaborating layers (www.calit2.net/research/layers.html). Research projects serve as vertical, multidisciplinary “convective currents” to draw and mix participation from across several layers. This brochure focuses on the Digitally Enabled Genomic Medicine (DeGeM, www.calit2.net/degem), the application layer of Cal-(IT)2 at UCSD, which is lead by John Wooley. Pierre Baldi is the layer leader of DeGeM for Cal-(IT)2 at partner institution UCI. Highlighting these selected projects and associated researchers will define the breadth and excitement of DeGeM and identify opportunities for involvement for interested academic researchers, students, and industrial partners. DeGeM: This acronym is pronounced “dee gem.” We have chosen to spell the abbreviation DeGeM to reflect the desired pronunciation.