Low GSTM3 Expression Is Associated with Poor Disease‐Free Survival In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

GST Polymorphisms and GST Enzyme Activity in Type 1 Myotonic Dystrophy
Journal of Investigative Genomics Research Article Open Access GST polymorphisms and GST enzyme activity in type 1 myotonic dystrophy Abstract Volume 2 Issue 2 - 2015 Glutathione S-transferases (GSTs) play an important role in protecting cells from damage Ashok Kumar,1 Sarita Agarwal,1 Suni,l caused by endogenous and exogenous compounds. In present study, we investigated the 2 association of GST gene polymorphisms (GSTT1, GSTM1, GSTP1 and GSTM3) and its Pradhan 1 enzyme activity in DM1 affected Indian population. Serum GST level was assessed by Department of Genetics, Sanjay Gandhi Post Graduate Institute of Medical Sciences, India using GST-kit. GSTM1 (null or present) & GSTT1 (null or present) GSTM3 (AA, AB 2Department of Neurology, Sanjay Gandhi Post Graduate and BB) & GSTP1 (Ile/Ile, Ile/Val and Val/Val -105) polymorphism were analyzed. The Institute of Medical Sciences, India serum GST significantly reduced in the patient group compared to the control group and significantly correlated with diabetes. Patients had significantly higher (except GSTM3A/B) Correspondence: Sarita Agarwal, Department of Genetics, GSTM1*0 (GSTM1 null genotype), GSTT1*0 and GSTP1 (ile/val) frequency than Sanjay Gandhi Post Graduate Institute of Medical Sciences, controls. The deletion frequencies (GSTM1 and GSTT1) and GSTP1 (ile/val) were not Lucknow, India- 226014, Tel 91 522 249 4349, Fax 9152 2266 associated with higher risk while heterozygous frequency of GSTM3 (A/B) increased risk 8017, Email [email protected] ratio up to three fold. The GSTM1, GSTT1, GSTM3 genotypes correlated with dyspepsia, age at presentation and duration of disease respectively. The allelic frequencies were 0.46, Received: January 12, 2015 | Published: March 04, 2015 0.54 and 0.48, 0.52 for GSTM3*A and GSTM3*B and were 0.61, 0.39 and 0.55, 0.45 for ile (A) and val (G) of GSTP1 for DM1 patients and control group respectively. -

Datasheet: VMA00551KT
Datasheet: VMA00551KT Description: GSTM3 ANTIBODY WITH CONTROL LYSATE Specificity: GSTM3 Format: Purified Product Type: PrecisionAb™ Monoclonal Clone: CPTC23 Isotype: IgG2a Quantity: 2 Westerns Product Details Applications This product has been reported to work in the following applications. This information is derived from testing within our laboratories, peer-reviewed publications or personal communications from the originators. Please refer to references indicated for further information. For general protocol recommendations, please visit www.bio-rad-antibodies.com/protocols. Yes No Not Determined Suggested Dilution Western Blotting 1/1000 PrecisionAb antibodies have been extensively validated for the western blot application. The antibody has been validated at the suggested dilution. Where this product has not been tested for use in a particular technique this does not necessarily exclude its use in such procedures. Further optimization may be required dependant on sample type. Target Species Human Species Cross Reacts with: Mouse Reactivity N.B. Antibody reactivity and working conditions may vary between species. Product Form Purified IgG - liquid Preparation 20μl Mouse monoclonal antibody purified by affinity chromatography on Protein G from tissue culture supernatant Buffer Solution Phosphate buffered saline Preservative 0.09% Sodium Azide (NaN ) Stabilisers 3 Immunogen Recombinant protein of human GSTM3 External Database Links UniProt: P21266 Related reagents Entrez Gene: 2947 GSTM3 Related reagents Page 1 of 3 Synonyms GST5 Specificity Mouse anti Human GSTM3 antibody recognizes the Glutathione S-transferase Mu 3, also known as GST class-mu 3, S-(hydroxyalkyl)glutathione lyase M3, brain GST, brain type mu-glutathione S-transferase or glutathione S-alkyltransferase M3. Cytosolic and membrane-bound forms of glutathione S-transferase are encoded by two distinct supergene families. -

Blood Biomarkers for Memory: Toward Early Detection of Risk for Alzheimer Disease, Pharmacogenomics, and Repurposed Drugs
Molecular Psychiatry (2020) 25:1651–1672 https://doi.org/10.1038/s41380-019-0602-2 IMMEDIATE COMMUNICATION Blood biomarkers for memory: toward early detection of risk for Alzheimer disease, pharmacogenomics, and repurposed drugs 1,2,3 1 1 1,3,4 1 1 1 3 A. B. Niculescu ● H. Le-Niculescu ● K. Roseberry ● S. Wang ● J. Hart ● A. Kaur ● H. Robertson ● T. Jones ● 3 3,5 5 2 1 1,4 4 A. Strasburger ● A. Williams ● S. M. Kurian ● B. Lamb ● A. Shekhar ● D. K. Lahiri ● A. J. Saykin Received: 25 March 2019 / Revised: 25 September 2019 / Accepted: 11 November 2019 / Published online: 2 December 2019 © The Author(s) 2019. This article is published with open access Abstract Short-term memory dysfunction is a key early feature of Alzheimer’s disease (AD). Psychiatric patients may be at higher risk for memory dysfunction and subsequent AD due to the negative effects of stress and depression on the brain. We carried out longitudinal within-subject studies in male and female psychiatric patients to discover blood gene expression biomarkers that track short term memory as measured by the retention measure in the Hopkins Verbal Learning Test. These biomarkers were subsequently prioritized with a convergent functional genomics approach using previous evidence in the field implicating them in AD. The top candidate biomarkers were then tested in an independent cohort for ability to predict state short-term memory, 1234567890();,: 1234567890();,: and trait future positive neuropsychological testing for cognitive impairment. The best overall evidence was for a series of new, as well as some previously known genes, which are now newly shown to have functional evidence in humans as blood biomarkers: RAB7A, NPC2, TGFB1, GAP43, ARSB, PER1, GUSB, and MAPT. -
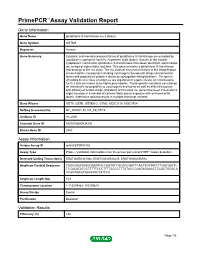
Download Validation Data
PrimePCR™Assay Validation Report Gene Information Gene Name glutathione S-transferase mu 3 (brain) Gene Symbol GSTM3 Organism Human Gene Summary Cytosolic and membrane-bound forms of glutathione S-transferase are encoded by two distinct supergene families. At present eight distinct classes of the soluble cytoplasmic mammalian glutathione S-transferases have been identified: alpha kappa mu omega pi sigma theta and zeta. This gene encodes a glutathione S-transferase that belongs to the mu class. The mu class of enzymes functions in the detoxification of electrophilic compounds including carcinogens therapeutic drugs environmental toxins and products of oxidative stress by conjugation with glutathione. The genes encoding the mu class of enzymes are organized in a gene cluster on chromosome 1p13.3 and are known to be highly polymorphic. These genetic variations can change an individual's susceptibility to carcinogens and toxins as well as affect the toxicity and efficacy of certain drugs. Mutations of this class mu gene have been linked with a slight increase in a number of cancers likely due to exposure with environmental toxins. Alternative splicing results in multiple transcript variants. Gene Aliases GST5, GSTB, GSTM3-3, GTM3, MGC3310, MGC3704 RefSeq Accession No. NC_000001.10, NT_032977.9 UniGene ID Hs.2006 Ensembl Gene ID ENSG00000134202 Entrez Gene ID 2947 Assay Information Unique Assay ID qHsaCEP0051802 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENST00000361066, -

Transcriptomic and Computational Analysis Identified LPA Metabolism
www.nature.com/scientificreports OPEN Transcriptomic and computational analysis identifed LPA metabolism, KLHL14 and KCNE3 as novel regulators of Epithelial- Mesenchymal Transition V. Di Lollo1,2,3*, A. Canciello 1,3*, M. Orsini2, N. Bernabò 1, M. Ancora2, M. Di Federico2, V. Curini2, M. Mattioli1, V. Russo1, A. Mauro1, C. Cammà2 & B. Barboni 1 Epithelial-mesenchymal transition (EMT) is a complex biological program between physiology and pathology. Here, amniotic epithelial cells (AEC) were used as in vitro model of transiently inducible EMT in order to evaluate the transcriptional insights underlying this process. Therefore, RNA-seq was used to identify the diferentially expressed genes and enrichment analyses were carried out to assess the intracellular pathways involved. As a result, molecules exclusively expressed in AEC that experienced EMT (GSTA1-1 and GSTM3) or when this process is inhibited (KLHL14 and KCNE3) were identifed. Lastly, the network theory was used to obtain a computational model able to recognize putative controller genes involved in the induction and in the prevention of EMT. The results suggested an opposite role of lysophosphatidic acid (LPA) synthesis and degradation enzymes in the regulation of EMT process. In conclusion, these molecules may represent novel EMT regulators and also targets for developing new therapeutic strategies. Epithelial-mesenchymal transition (EMT) is a complex biological process. Although intracellular pathways lead- ing to the trans-diferentiation of epithelial into fbroblastic-like cells are known, several molecular mechanisms still remain to be clarifed despite the clinical urgency of the issue1. At the present, EMT is recognized to play an essential role in driving three diferent biological events: (1) type 1 EMT during embryo development, (2) type 2 in the fbrotic process, wound healing and tissue regeneration while (3) type 3 during metastatic process2. -

Role of GSTM3 Polymorphism in the Risk of Developing Esophageal Cancer
178 Short Communication Role of GSTM3 Polymorphism in the Risk of Developing Esophageal Cancer Meenu Jain,1 Shaleen Kumar,2 Punita Lal,2 Anu Tiwari,2 Uday C. Ghoshal,3 and Balraj Mittal1 Departments of 1Medical Genetics, 2Radiotherapy, and 3Gastroenterology, Sanjay Gandhi Post Graduate Institute of Medical Sciences, Lucknow, India Abstract GSTM3 is involved in detoxification of carcinogens and may GSTM3 AB + BB and GSTM1 null genotypes marginally be important in modulating cancer susceptibility. GSTM3 modulated risk (OR, 2.3; 95% CI, 1.1-3.7; P = 0.01). Association genotype frequencies were determined in peripheral blood with histology (adenocarcinoma: OR, 3.4; 95% CI, 1.1-10.9; DNA of 149 esophageal cancer patients and 200 nonmalignant P = 0.03) and tumor site (middle third location: OR, 2.2; 95% controls using the PCR followed by PAGE. Patients who were CI, 1.1-4.4; P = 0.01; lower third location: OR, 2.6; 95% CI, 1.2- heterozygous carriers of GSTM3 AB genotype had an 5.6; P = 0.01) was also documented. Our results suggest that enhanced risk for developing esophageal cancer [odds ratio GSTM3 polymorphism may influence esophageal cancer (OR), 2.1; 95% confidence interval (95% CI), 1.1-3.7; P = 0.01]. susceptibility, in particular modulating the risk for adeno- In males, the risk due to GSTM3 AB genotype increased carcinoma histology and tumors of the mid and lower third further (OR, 3.4; 95% CI, 1.7-6.8; P = 0.000). Interaction of region. (Cancer Epidemiol Biomarkers Prev 2007;16(1):178–81) Introduction Nutritional deficiency coupled with alcohol and tobacco abuse mutation, the expression of GSTM3 can be influenced. -

Glutathione S Transferases
ooggeenneessii iinn ss && rrcc aa MM CC uu tt ff aa Journal ofJournal of oo gg ll ee aa nn nn ee rr ss uu ii Nissar et al., J Carcinog Mutagen 2017, 8:2 ss oo JJ ISSN: 2157-2518 CarCarcinogenesiscinogenesis & Mutagenesis DOI: 10.4172/2157-2518.1000287 Review Article Open Access Glutathione S Transferases: Biochemistry, Polymorphism and Role in Colorectal Carcinogenesis Saniya Nissar1,2,3, Aga Syed Sameer4*, Roohi Rasool2, Nissar A Chowdri5 and Fouzia Rashid3 1Department of Biochemistry, University of Kashmir, India 2Department of Immunology and Molecular Medicine, Sher-I-Kashmir Institute of Medical Sciences, India 3Department of Clinical Biochemistry, University of Kashmir, India 4Department of Basic Medical Sciences, College of Medicine, King Saud Bin Abdulaziz University for Health Sciences. Jeddah, KSA 5Department of General Surgery, Sher-I-Kashmir Institute of Medical Sciences, India *Corresponding author: Aga Syed Sameer, Department of Basic Medical Sciences, College of Medicine, King Saud Bin Abdulaziz University for Health Sciences. Jeddah, KSA, Tel: +966542035086; E-mail: [email protected] Received date: February 19, 2017; Accepted date: March 10, 2017; Published date: March 15, 2017 Copyright: © 2017 Nissar S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Abstract Glutathione S-transferases (GSTs) are enzymes detoxifying a wide range of hazardous substances both of endogenous or exogenous origin, such as reactive oxygen species (ROS) or xenobiotics and environmental carcinogens; thereby imparting protection to DNA against oxidative damage. -

GSTM3 (NM 000849) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC201013 GSTM3 (NM_000849) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: GSTM3 (NM_000849) Human Tagged ORF Clone Tag: Myc-DDK Symbol: GSTM3 Synonyms: GST5; GSTB; GSTM3-3; GTM3 Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin ORF Nucleotide >RC201013 representing NM_000849 Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGTCGTGCGAGTCGTCTATGGTTCTCGGGTACTGGGATATTCGTGGGCTGGCGCACGCCATCCGCCTGC TCCTGGAGTTCACGGATACCTCTTATGAGGAGAAACGGTACACGTGCGGGGAAGCTCCTGACTATGATCG AAGCCAATGGCTGGATGTGAAATTCAAGCTAGACCTGGACTTTCCTAATCTGCCCTACCTCCTGGATGGG AAGAACAAGATCACCCAGAGCAATGCCATCTTGCGCTACATCGCTCGCAAGCACAACATGTGTGGTGAGA CTGAAGAAGAAAAGATTCGAGTGGACATCATAGAGAACCAAGTAATGGATTTCCGCACACAACTGATAAG GCTCTGTTACAGCTCTGACCACGAAAAACTGAAGCCTCAGTACTTGGAAGAGCTACCTGGACAACTGAAA CAATTCTCCATGTTTCTGGGGAAATTCTCATGGTTTGCCGGGGAAAAGCTCACCTTTGTGGATTTTCTCA CCTATGATATCTTGGATCAGAACCGTATATTTGACCCCAAGTGCCTGGATGAGTTCCCAAACCTGAAGGC TTTCATGTGCCGTTTTGAGGCTTTGGAGAAAATCGCTGCCTACTTACAGTCTGATCAGTTCTGCAAGATG CCCATCAACAACAAGATGGCCCAGTGGGGCAACAAGCCTGTATGC ACGCGTACGCGGCCGCTCGAGCAGAAACTCATCTCAGAAGAGGATCTGGCAGCAAATGATATCCTGGATT ACAAGGATGACGACGATAAGGTTTAA This product is to be used for laboratory only. Not for diagnostic or therapeutic -

Phase I and II Carcinogen Metabolism Gene Expression in Human Lung Tissue and Tumors
6002 Vol. 9, 6002–6011, December 1, 2003 Clinical Cancer Research Phase I and II Carcinogen Metabolism Gene Expression in Human Lung Tissue and Tumors Simon D. Spivack,1 Gregory J. Hurteau,2 and gender factors, as well as ongoing tobacco exposure. Michael J. Fasco,1 and Laurence S. Kaminsky1 Expression differences between tumor and nontumor tissue in this pathway have both susceptibility and therapeutic 1Wadsworth Center, New York State Department of Health, Albany, New York, and 2University of Colorado Health Science Center, implications. Denver, Colorado INTRODUCTION One in 10 smokers develops lung cancer over a lifetime. ABSTRACT Nonrandom clustering of cases in families indicates that some of Purpose: The regulation of carcinogen metabolism ma- this proclivity is inherited (1–9), possibly through low-pen- chinery may involve proximate tobacco smoke exposure, etrance, multigene loci (10). Because mainstream tobacco hormonal and other endogenous coregulatory factors, and smoke is the primary cause of lung cancer, the carcinogen an individual’s underlying genetic responsiveness. The metabolism genes comprise a plausible lung cancer susceptibil- mRNA and protein expression patterns of known carcino- ity pathway (1, 11). gen metabolism genes encoding the aromatic hydrocarbon Tobacco exposure measurement by self-reported smoking receptor Ahr; the cytochromes P450 CYP1A1 and CYP1B1; history can cause misclassification bias; an alternative approach glutathione S-transferases GSTM1, GSTM3, GSTP1, and has been to measure the exposure biomarker nicotine and its GSTT1; and NADPH quinone oxidoreductase NQO1 were metabolite cotinine (12). Nicotine has a plasma half-life of ϳ2 examined. h, whereas the half-life of plasma cotinine is ϳ17 h (13). -
GSTM3 Antibody Cat
GSTM3 Antibody Cat. No.: 30-882 GSTM3 Antibody Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Human Antibody produced in rabbits immunized with a synthetic peptide corresponding a region IMMUNOGEN: of human GSTM3. TESTED APPLICATIONS: ELISA, WB GSTM3 antibody can be used for detection of GSTM3 by ELISA at 1:312500. GSTM3 APPLICATIONS: antibody can be used for detection of GSTM3 by western blot at 1 μg/mL, and HRP conjugated secondary antibody should be diluted 1:50,000 - 100,000. POSITIVE CONTROL: 1) Cat. No. 1211 - HepG2 Cell Lysate PREDICTED MOLECULAR 26 kDa WEIGHT: Properties PURIFICATION: Antibody is purified by peptide affinity chromatography method. CLONALITY: Polyclonal CONJUGATE: Unconjugated PHYSICAL STATE: Liquid September 26, 2021 1 https://www.prosci-inc.com/gstm3-antibody-30-882.html Purified antibody supplied in 1x PBS buffer with 0.09% (w/v) sodium azide and 2% BUFFER: sucrose. CONCENTRATION: batch dependent For short periods of storage (days) store at 4˚C. For longer periods of storage, store STORAGE CONDITIONS: GSTM3 antibody at -20˚C. As with any antibody avoid repeat freeze-thaw cycles. Additional Info OFFICIAL SYMBOL: GSTM3 ALTERNATE NAMES: GSTM3, GST5, GSTB, GSTM3-3, GTM3, MGC3310, MGC3704 ACCESSION NO.: NP_000840 PROTEIN GI NO.: 23065552 GENE ID: 2947 USER NOTE: Optimal dilutions for each application to be determined by the researcher. Background and References Cytosolic and membrane-bound forms of glutathione S-transferase are encoded by two distinct supergene families. At present, eight distinct classes of the soluble cytoplasmic mammalian glutathione S-transferases have been identified: alpha, kappa, mu, omega, pi, sigma, theta and zeta. GSTM3 is a glutathione S-transferase that belongs to the mu class. -

Expression-Based Discovery of Variation in the Human Glutathione
Research Article Expression-Based Discovery of Variation in the Human Glutathione S-Transferase M3 Promoter and Functional Analysis in a Glioma Cell Line Using Allele-Specific Chromatin Immunoprecipitation Xuemei Liu, Michelle R. Campbell, Gary S. Pittman, Eric C. Faulkner, Mary A. Watson, and Douglas A. Bell Laboratory of Computational Biology and Risk Analysis, National Institute of Environmental Health Sciences, NIH, Research Triangle Park, North Carolina Abstract on the efficacy of therapeutic treatments (16), characterization of Discovery and functional evaluation of biologically significant genotypic and phenotypic variation in the GST gene family is regulatory single nucleotide polymorphisms (SNP) in carcin- important. ogen metabolism genes is a difficult challenge because the GSTM3 is expressed in testis, brain, lung, and lymphocytes as phenotypic consequences may be both transient and subtle. well as in many tumor types (NCI-60; refs. 17–19). A recently We have used a gene expression screening approach to identify described GSTM3 3-bp deletion polymorphism (GSTM3*A/*B) has a functional regulatory SNP in glutathione S-transferase M3 been shown to have a frequency of 0.16 in the Caucasian population (GSTM3). Anttila et al. proposed that variation in GSTM3 (20, 21). This polymorphism was associated with an increased risk expression was affected by exposure to cigarette smoke and of cutaneous basal cell carcinomas and laryngeal carcinomas and inheritance of the GSTM1-null genotype. To investigate the interacted with other GST variants in modifying astrocytoma risk but had no effect on pharyngeal carcinoma risk (20–24). Anttila mechanism of GSTM3 expression variation, we measured GSTM3 expression in lymphoblast cells from a human Centre et al. -

Role of GSTM3 Polymorphism in the Risk of Developing Esophageal Cancer
178 Short Communication Role of GSTM3 Polymorphism in the Risk of Developing Esophageal Cancer Meenu Jain,1 Shaleen Kumar,2 Punita Lal,2 Anu Tiwari,2 Uday C. Ghoshal,3 and Balraj Mittal1 Departments of 1Medical Genetics, 2Radiotherapy, and 3Gastroenterology, Sanjay Gandhi Post Graduate Institute of Medical Sciences, Lucknow, India Abstract GSTM3 is involved in detoxification of carcinogens and may GSTM3 AB + BB and GSTM1 null genotypes marginally be important in modulating cancer susceptibility. GSTM3 modulated risk (OR, 2.3; 95% CI, 1.1-3.7; P = 0.01). Association genotype frequencies were determined in peripheral blood with histology (adenocarcinoma: OR, 3.4; 95% CI, 1.1-10.9; DNA of 149 esophageal cancer patients and 200 nonmalignant P = 0.03) and tumor site (middle third location: OR, 2.2; 95% controls using the PCR followed by PAGE. Patients who were CI, 1.1-4.4; P = 0.01; lower third location: OR, 2.6; 95% CI, 1.2- heterozygous carriers of GSTM3 AB genotype had an 5.6; P = 0.01) was also documented. Our results suggest that enhanced risk for developing esophageal cancer [odds ratio GSTM3 polymorphism may influence esophageal cancer (OR), 2.1; 95% confidence interval (95% CI), 1.1-3.7; P = 0.01]. susceptibility, in particular modulating the risk for adeno- In males, the risk due to GSTM3 AB genotype increased carcinoma histology and tumors of the mid and lower third further (OR, 3.4; 95% CI, 1.7-6.8; P = 0.000). Interaction of region. (Cancer Epidemiol Biomarkers Prev 2007;16(1):178–81) Introduction Nutritional deficiency coupled with alcohol and tobacco abuse mutation, the expression of GSTM3 can be influenced.