Lathyrus Sativus) for Low Β-L-ODAP Content
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Multiple Polyploidy Events in the Early Radiation of Nodulating And
Multiple Polyploidy Events in the Early Radiation of Nodulating and Nonnodulating Legumes Steven B. Cannon,*,y,1 Michael R. McKain,y,2,3 Alex Harkess,y,2 Matthew N. Nelson,4,5 Sudhansu Dash,6 Michael K. Deyholos,7 Yanhui Peng,8 Blake Joyce,8 Charles N. Stewart Jr,8 Megan Rolf,3 Toni Kutchan,3 Xuemei Tan,9 Cui Chen,9 Yong Zhang,9 Eric Carpenter,7 Gane Ka-Shu Wong,7,9,10 Jeff J. Doyle,11 and Jim Leebens-Mack2 1USDA-Agricultural Research Service, Corn Insects and Crop Genetics Research Unit, Ames, IA 2Department of Plant Biology, University of Georgia 3Donald Danforth Plant Sciences Center, St Louis, MO 4The UWA Institute of Agriculture, The University of Western Australia, Crawley, WA, Australia 5The School of Plant Biology, The University of Western Australia, Crawley, WA, Australia 6Virtual Reality Application Center, Iowa State University 7Department of Biological Sciences, University of Alberta, Edmonton, AB, Canada 8Department of Plant Sciences, The University of Tennessee Downloaded from 9BGI-Shenzhen, Bei Shan Industrial Zone, Shenzhen, China 10Department of Medicine, University of Alberta, Edmonton, AB, Canada 11L. H. Bailey Hortorium, Department of Plant Biology, Cornell University yThese authors contributed equally to this work. *Corresponding author: E-mail: [email protected]. http://mbe.oxfordjournals.org/ Associate editor:BrandonGaut Abstract Unresolved questions about evolution of the large and diverselegumefamilyincludethetiming of polyploidy (whole- genome duplication; WGDs) relative to the origin of the major lineages within the Fabaceae and to the origin of symbiotic nitrogen fixation. Previous work has established that a WGD affects most lineages in the Papilionoideae and occurred sometime after the divergence of the papilionoid and mimosoid clades, but the exact timing has been unknown. -
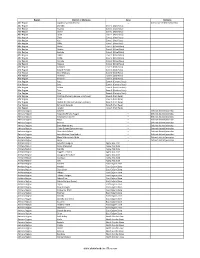
Districts of Ethiopia
Region District or Woredas Zone Remarks Afar Region Argobba Special Woreda -- Independent district/woredas Afar Region Afambo Zone 1 (Awsi Rasu) Afar Region Asayita Zone 1 (Awsi Rasu) Afar Region Chifra Zone 1 (Awsi Rasu) Afar Region Dubti Zone 1 (Awsi Rasu) Afar Region Elidar Zone 1 (Awsi Rasu) Afar Region Kori Zone 1 (Awsi Rasu) Afar Region Mille Zone 1 (Awsi Rasu) Afar Region Abala Zone 2 (Kilbet Rasu) Afar Region Afdera Zone 2 (Kilbet Rasu) Afar Region Berhale Zone 2 (Kilbet Rasu) Afar Region Dallol Zone 2 (Kilbet Rasu) Afar Region Erebti Zone 2 (Kilbet Rasu) Afar Region Koneba Zone 2 (Kilbet Rasu) Afar Region Megale Zone 2 (Kilbet Rasu) Afar Region Amibara Zone 3 (Gabi Rasu) Afar Region Awash Fentale Zone 3 (Gabi Rasu) Afar Region Bure Mudaytu Zone 3 (Gabi Rasu) Afar Region Dulecha Zone 3 (Gabi Rasu) Afar Region Gewane Zone 3 (Gabi Rasu) Afar Region Aura Zone 4 (Fantena Rasu) Afar Region Ewa Zone 4 (Fantena Rasu) Afar Region Gulina Zone 4 (Fantena Rasu) Afar Region Teru Zone 4 (Fantena Rasu) Afar Region Yalo Zone 4 (Fantena Rasu) Afar Region Dalifage (formerly known as Artuma) Zone 5 (Hari Rasu) Afar Region Dewe Zone 5 (Hari Rasu) Afar Region Hadele Ele (formerly known as Fursi) Zone 5 (Hari Rasu) Afar Region Simurobi Gele'alo Zone 5 (Hari Rasu) Afar Region Telalak Zone 5 (Hari Rasu) Amhara Region Achefer -- Defunct district/woredas Amhara Region Angolalla Terana Asagirt -- Defunct district/woredas Amhara Region Artuma Fursina Jile -- Defunct district/woredas Amhara Region Banja -- Defunct district/woredas Amhara Region Belessa -- -

Report on the 8Th International Workshop on the CCN Family of Genes – Nice November 3–8, 2015
J. Cell Commun. Signal. (2016) 10:77–86 DOI 10.1007/s12079-016-0317-y EDITORIAL Report on the 8th international workshop on the CCN family of genes – Nice November 3–8, 2015 Bernard Perbal1 & Lester Lau 2 & Karen Lyons3 & Satoshi Kubota4 & Herman Yeger5 & Gary Fisher6 Received: 29 January 2016 /Accepted: 29 January 2016 /Published online: 26 February 2016 # The International CCN Society 2016 The 8th international workshop on the CCN family of genes was able and enthusiastic about participating with us in the full was held for a second time in Nice, from November 3rd to 8th, sessions of the meeting. It is always a unique and fruitful 2015. Under particularly sunny and warm weather everyone opportunity for all participants, either young or established enjoyed the charms of the Mediterranean coastline. scientists, to share time with the awardees. Thanks to Annick Perbal, the social events during the meet- This year we also had the pleasure to welcome Robert ing were a real success and were truly well appreciated by all. Baxter, a former ICCNS-Springer awardee, who flew in from The choice of the Westminster hotel was an excellent one. Australia to join us and present a special conference on the role of This year the ICCNS honored and warmly welcomed IGFBP3 in the breast cancer response to DNA damaging Judith Campisi, the recipient of the 2015 ICCNS-Springer therapy. award, who accepted to present a conference on senescence Those who did not have the opportunity to make it at the time and cancer. We are grateful to Judith Campisi who came a Robert Baxter received the award in Sydney, could then enjoy long way from San Francisco, despite being under the weather sharing experiences and discuss possible levels of interactions with a bad cold and sore throat. -

Ecogeographic, Genetic and Taxonomic Studies of the Genus Lathyrus L
ECOGEOGRAPHIC, GENETIC AND TAXONOMIC STUDIES OF THE GENUS LATHYRUS L. BY ALI ABDULLAH SHEHADEH A thesis submitted to the University of Birmingham for the degree of DOCTOR OF PHILOSOPHY School of Biosciences College of Life and Environmental Sciences University of Birmingham March 2011 University of Birmingham Research Archive e-theses repository This unpublished thesis/dissertation is copyright of the author and/or third parties. The intellectual property rights of the author or third parties in respect of this work are as defined by The Copyright Designs and Patents Act 1988 or as modified by any successor legislation. Any use made of information contained in this thesis/dissertation must be in accordance with that legislation and must be properly acknowledged. Further distribution or reproduction in any format is prohibited without the permission of the copyright holder. ABSTRACT Lathyrus species are well placed to meet the increasing global demand for food and animal feed, at the time of climate change. Conservation and sustainable use of the genetic resources of Lathyrus is of significant importance to allow the regain of interest in Lathyrus species in world. A comprehensive global database of Lathyrus species originating from the Mediterranean Basin, Caucasus, Central and West Asia Regions is developed using accessions in major genebanks and information from eight herbaria in Europe. This Global Lathyrus database was used to conduct gap analysis to guide future collecting missions and in situ conservation efforts for 37 priority species. The results showed the highest concentration of Lathyrus priority species in the countries of the Fertile Crescent, France, Italy and Greece. -

Nutrition Surveys 1999
NUTRITION SURVEYS 1999 – 2000 Region/Zone woreda Date of Agency Sample size Methodology Nutrition Indicatorsi survey Tigre throughout August SCF-UK 937 30 cluster Mean WHL <80%WFL W/H<-2 Z score W/H <-3 Z score 1999 92.8% 5.5% 7.7 % 1.0% Tigre Feb 2000 WVI W/H <-2 Z score W/H <-3 Z score Eastern May 2000 Feb 2000 May 2000 Feb 2000 May 2000 Asti Wenberta 685 13.1% 10.9% NA 2.6% Saesi Tsaedaemba 1412 22.3% 20.1% 3.7% 4.4% Amhara: May-June SCF-UK + 2900 58 clusters in Mean WFL < 80% WFL N.Wello Bugna 1999 worst drought 88.8% 4% Wadla affected woredas. 89,4% 7% Gidan 87.8% 3% Delanta Dawnt 89.4% 7% Gubalafto 90.0% 7% S. Wello Dessie Zuria 89.8% 7% Tenta 90.5% 3% Legambo 90.8% 5% Ambassel 90.7% 6% Mekdella 91.2% 4% Wag Hamra Dehana 88.2% 4% Oromyia Chefa 92.8% 2% Amhara Aug - Oct SCF-UK + 2500 50 clusters in Mean WFL < 80% WFL 1999 worst drought Aug Sept Oct Aug Sept Oct N.Wello Bugna affected woredas. 91.2% 88.7% 89.7% 6.6% 10.6% 7.0% Wadla 91.1% 90.7% 90.6% 7.3% 5.5% 5.7% Gidan 88.4% 88.2% 88.4% 8.9% 8.2% 9.6% S. Wello Delanta Dawnt 87.5% 87.6% 87.5% 11.0% 8.0% 7.6% Dessie Zuria 91.9% 90.9% 90.1% 4.0% 6.2% 5.2% Tenta 89.2% 89.1% 88.4% 10.0% 8.3% 11.7% Legambo 89.1% 89.7% 89.6% 8.4% 6.0% 6.4% Wag Hamra Dehana 89.7% 89.5% 90.5% 8.3% 8.2% 6.4% Amhara March- May SCF-UK + 2500 50 clusters in Mean WFL < 80% WFL N.Wello 2000 worst drought March 2000 May 2000 March 2000 May 2000 Bugna affected woredas 90.1% 90.6% Wadla 90.5% 91.1% S. -

Ethnobotany, Diverse Food Uses, Claimed Health Benefits And
Shewayrga and Sopade Journal of Ethnobiology and Ethnomedicine 2011, 7:19 http://www.ethnobiomed.com/content/7/1/19 JOURNAL OF ETHNOBIOLOGY AND ETHNOMEDICINE RESEARCH Open Access Ethnobotany, diverse food uses, claimed health benefits and implications on conservation of barley landraces in North Eastern Ethiopia highlands Hailemichael Shewayrga1* and Peter A Sopade2,3 Abstract Background: Barley is the number one food crop in the highland parts of North Eastern Ethiopia produced by subsistence farmers grown as landraces. Information on the ethnobotany, food utilization and maintenance of barley landraces is valuable to design and plan germplasm conservation strategies as well as to improve food utilization of barley. Methods: A study, involving field visits and household interviews, was conducted in three administrative zones. Eleven districts from the three zones, five kebeles in each district and five households from each kebele were visited to gather information on the ethnobotany, the utilization of barley and how barley end-uses influence the maintenance of landrace diversity. Results: According to farmers, barley is the “king of crops” and it is put for diverse uses with more than 20 types of barley dishes and beverages reportedly prepared in the study area. The products are prepared from either boiled/roasted whole grain, raw- and roasted-milled grain, or cracked grain as main, side, ceremonial, and recuperating dishes. The various barley traditional foods have perceived qualities and health benefits by the farmers. Fifteen diverse barley landraces were reported by farmers, and the ethnobotany of the landraces reflects key quantitative and qualitative traits. Some landraces that are preferred for their culinary qualities are being marginalized due to moisture shortage and soil degradation. -

British Geological Survey PLANNING
British Geological Survey TECHNICAL REPORT WC/00/13 Overseas Geology Series PLANNING FOR GROUNDWATER DROUGHT IN AFRICA: TOWARDS A SYSTEMATIC APPROACH FOR ASSESSING WATER SECURITY IN ETHIOPIA Project report on visit to Ethiopia, November-December 1999 British Geological Survey, Wallingford British Geological Survey TECHNICAL REPORT WC/00/13 Overseas Geology Series PLANNING FOR GROUNDWATER DROUGHT IN AFRICA: TOWARDS A SYSTEMATIC APPROACH FOR ASSESSING WATER SECURITY IN ETHIOPIA Project report on visit to Ethiopia, November-December 1999 R C Calow1, A M MacDonald1 and A L Nicol2 1British Geological Survey 2Overseas Development Institute This document is an output from a project funded by the Department for International Development (DFID) for the benefit of developing countries. The views expressed are not necessarily those of the DFID. DFID classification: Subsector: Water and Sanitation Theme: W4 – Raise the well-being of the rural and urban poor through cost-effective improved water supply and sanitation Project Title: Groundwater drought early warning for vulnerable areas Project reference: R7125 Bibliographic reference: Calow, R C, MacDonald, A M and Nicol, A L 2000. Planning for Groundwater Drought in Africa. Towards a Systematic Approach for Assessing Water Security in Ethiopia. Project report on visit to Ethiopia, November – December 1999 BGS Technical Report WC/00/13 Keywords:Ethiopia, groundwater, drought, monitoring Front cover illustration: Looking down onto the River Mille from Abbot Village, Ambassel Woreda. © NERC 2000 Keyworth, Nottingham, British Geological Survey, 2000 Contents Executive Summary iv 1. INTRODUCTION 1 1.1 Visit Objectives 1 1.2 Visit Background 1 1.3 Project Aims and Outputs 3 2. THE STUDY AREA: SOUTH WOLLO 4 2.1 Selection Criteria 4 2.2 Physical Background 5 2.3 Socio-economic Background 5 2.4 Institutions 6 3. -

Variation in the Grass Pea (Lathyrus Sa Tivus L.' and Wild Species
Euphytica 33 (1984) 549-559 VARIATION IN THE GRASS PEA (LATHYRUS SA TIVUS L.' AND WILD SPECIES M. T. JACKSON and A. G. YUNUS1 Department of Plant Biology, University of Birmingham, Bl5 2Tr, England Received 21 September 1983 .DEX WORDS Lathyrus sativus,grass pea, wild species,variation, multivariate analyses. SUMMARY Forty-nine accessionsof Lathyrus sativuswere studied for morphological variation. Data were analysed using Principal Components Analysis and Cluster Analysis. The variation in 14 speciesof SectionLathyrus was also evaluated in order to ascertainaffinities betweenL. sativusand other species. L. sativus is a highly variable species,and there is a clear distinction betweenthe blue-flowered fonns from south-west Asia, Ethiopia and the Indian subcontinent, and the white and white and blue flowered fonns with white seedswhich have a more westerly distribution. Differences in vegetative parts may be due to selectionfor forage types. L. sativus appears to be closely related to L. cicera and L. gorgoni, and this relationship needs further investigation. INTRODUCTION The genus Lathyrus is large with 187 speciesand subspeciesrecognised (ALLKIN et al., 1983).Species are found in the Old World and the New World, but clearly there are centres of diversity for Old World speciesin Asia Minor and the Mediterranean region (ZEVEN& DE WET,1982). A number of speciesare usedas animal fodder plants including L. hirsutusand L. palustris, and some are valued as ornamentals, especially L. odoratus,the sweetpea. Only one species,L. sativus,the grasspea, khesari or chick- ling pea is widely cultivated as a food crop, and this pulse is a dependable cropper in drought conditions (SMARTT,1976). -

S P E C I a L R E P O
S P E C I A L R E P O R T FAO/WFP CROP AND FOOD SECURITY ASSESSMENT MISSION TO ETHIOPIA (Phase 2) Integrating the Crop and Food Supply and the Emergency Food Security Assessments 27 July 2009 FOOD AND AGRICULTURE ORGANIZATION OF THE UNITED NATIONS, ROME WORLD FOOD PROGRAMME, ROME - 2 - This report has been prepared by Mario Zappacosta, Jonathan Pound and Prisca Kathuku, under the responsibility of the FAO and WFP Secretariats. It is based on information from official and other sources. Since conditions may change rapidly, please contact the undersigned if further information is required. Henri Josserand Mustapha Darboe Deputy Director, GIEWS, FAO Regional Director for Southern, Eastern Fax: 0039-06-5705-4495 and Central Africa, WFP E-mail: [email protected] Fax: 0027-11-5171634 E-mail: : [email protected] Please note that this Special Report is also available on the Internet as part of the FAO World Wide Web (www.fao.org ) at the following URL address: http://www.fao.org/giews/ The Special Alerts/Reports can also be received automatically by E-mail as soon as they are published, by subscribing to the GIEWS/Alerts report ListServ. To do so, please send an E-mail to the FAO-Mail-Server at the following address: [email protected] , leaving the subject blank, with the following message: subscribe GIEWSAlertsWorld-L To be deleted from the list, send the message: unsubscribe GIEWSAlertsWorld-L Please note that it is now possible to subscribe to regional lists to only receive Special Reports/Alerts by region: Africa, Asia, Europe or Latin America (GIEWSAlertsAfrica-L, GIEWSAlertsAsia-L, GIEWSAlertsEurope-L and GIEWSAlertsLA-L). -

Neurotoxicity: Identifying and Controlling Poisons of the Nervous System
Index -i abused drugs activities of, 33, 81, 88, 322 addiction, 6, 52,74, 75 aldicarb, 47, 173,251 designer drugs, 51 aldrin, 250,252,292 effects on nervous system, 6,9, 51–53, 71 allyl chloride, 303 health costs of, 20,53,232 aluminum, 48 psychoactive drugs, 6, 10,27,44,50 and Alzheimer’s disease, 54-55 withdrawal from, 74 oxide, 14, 136 see also specific drugs Alzheimer’s disease academic research effects on hippocampus, 71 cooperative agreements with government, 94 environmental cause, 3, 6, 54-55, 70, 72 factors influencing directions of, 92-94 research on, 259 acetaldehyde, 297 American Academy of Pediatrics, 189 acetone, 14, 136,296, 297,303 American Conference of Governmental Industrial Hygienists, acetylcholine, 26, 66, 109, 124,294, 336 28, 151, 185, 186,203,302-303 acetylcholinesterase, 11,50,74,84,187,203, 289,290-292,336 American National Standards Institute, 185 acetylethyl tetramethyl tetralin (AETT) ammonia, 14, 136 exposure route, 108 amphetamines, 74 incidents of poisoning, 47, 54 amyotrophic lateral sclerosis neurological effects of, 54 characteristics of, 54 acrylamide, 73, 120 environmental cause, 3, 6, 54-55, 70, 72 neurotoxicity testing, 166, 175 incidence of, 54, 55 regulation for neurotoxicity, 178 research on, 259 risk assessment approaches, 150, 151,216 anencephaly, 70 research on neurotoxicity, 258 anilines and substituted anilines, 179 acrylates, 175 animal tests, 13 acrylonitrile, 203 accuracy and reliability, 106, 115 neurotoxicity testing, 166, 173 advantages and limitations of, 105, 106, 111, 112, 114, -
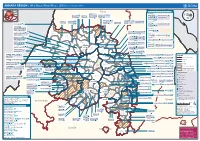
AMHARA REGION : Who Does What Where (3W) (As of 13 February 2013)
AMHARA REGION : Who Does What Where (3W) (as of 13 February 2013) Tigray Tigray Interventions/Projects at Woreda Level Afar Amhara ERCS: Lay Gayint: Beneshangul Gumu / Dire Dawa Plan Int.: Addis Ababa Hareri Save the fk Save the Save the df d/k/ CARE:f k Save the Children:f Gambela Save the Oromia Children: Children:f Children: Somali FHI: Welthungerhilfe: SNNPR j j Children:l lf/k / Oxfam GB:af ACF: ACF: Save the Save the af/k af/k Save the df Save the Save the Tach Gayint: Children:f Children: Children:fj Children:l Children: l FHI:l/k MSF Holand:f/ ! kj CARE: k Save the Children:f ! FHI:lf/k Oxfam GB: a Tselemt Save the Childrenf: j Addi Dessie Zuria: WVE: Arekay dlfk Tsegede ! Beyeda Concern:î l/ Mirab ! Concern:/ Welthungerhilfe:k Save the Children: Armacho f/k Debark Save the Children:fj Kelela: Welthungerhilfe: ! / Tach Abergele CRS: ak Save the Children:fj ! Armacho ! FHI: Save the l/k Save thef Dabat Janamora Legambo: Children:dfkj Children: ! Plan Int.:d/ j WVE: Concern: GOAL: Save the Children: dlfk Sahla k/ a / f ! ! Save the ! Lay Metema North Ziquala Children:fkj Armacho Wegera ACF: Save the Children: Tenta: ! k f Gonder ! Wag WVE: Plan Int.: / Concern: Save the dlfk Himra d k/ a WVE: ! Children: f Sekota GOAL: dlf Save the Children: Concern: Save the / ! Save: f/k Chilga ! a/ j East Children:f West ! Belesa FHI:l Save the Children:/ /k ! Gonder Belesa Dehana ! CRS: Welthungerhilfe:/ Dembia Zuria ! î Save thedf Gaz GOAL: Children: Quara ! / j CARE: WVE: Gibla ! l ! Save the Children: Welthungerhilfe: k d k/ Takusa dlfj k -

The Case of Dessie Zuria Woreda
CORE Metadata, citation and similar papers at core.ac.uk Provided by International Institute for Science, Technology and Education (IISTE): E-Journals Journal of Economics and Sustainable Development www.iiste.org ISSN 2222-1700 (Paper) ISSN 2222-2855 (Online) DOI: 10.7176/JESD Vol.10, No.5, 2019 Determinants of Households Saving Capacity and Bank Account Holding Experience in Ethiopia: The Case of Dessie Zuria Woreda Bazezew Endalew College of Business and Economics, Department of Economics, Wollo University, Dessie, Ethiopia Abstract This research has been an attempt to identify the major determinants that affect households saving capacity and their experience of adopting formal financial institutions (banks) in the case of Dessie Zuria Woreda. To do so, an individual base cross-sectional data analysis along with the two stage sampling technique of both purposive and random sampling technique was undertaken. To analyze the data, the study employed two sets of models (logistic and the method of principal component analysis). The econometric results of the study indicates that determinants like lack of credit access, lack of financial planning, complexity of banking system, monthly expenditure on stimulants, sex, significantly and negatively affects households saving capacity, but monthly income, age, bank account holding experience, marital status, and occupation positively and significantly affects saving capacity. In similar fashion, determinants include improper government policy, weak institutional set up, complexity of banking system, distance in Km away from their home to financial institutions, and religion significantly and negatively affect the probability of households to be banked, on the other hand, sex of households, credit access, income, marital status, education and age positively and significantly affects the probability of households to be banked.