Shining the Light on the Structural Color of Tenacibaculum Ectocooler
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diffractive Properties of Blue Morpho Butterfly Wings
Diffractive Properties of Blue Morpho Butterfly Wings Mary Lalak and Paul Brackman Department of Physics and Astronomy, University of Georgia, Athens, Georgia 30602 (Dated: October 24, 2014) Many species of butterflies are known to produce beautiful iridescent colors when exposed to light from different angles. These affects can be attributed to a few different optical phenomena combined, the most prominent being reflective diffraction. Using common household items and with a budget of 50 dollars, we attempt to confirm the theory that the collection of ridges on the individual scales on the wing act as transmission gratings, as well as reflective. These results are quantified by the calculation of the ridge separation and comparing this distance to those observed by a scanning electron microscope photo. I. INTRODUCTION In studying the optical property of iridescence, three distinct mechanisms must be discussed. Thin film inter- ference, structured coloring, and reflective diffraction all contribute to the iridescent qualities of a surface. Thin film interference occurs when light strikes a film, some of it enters the film, and the rest is reflected off. The light transmitted through reflects off the bottom of the film and exits the film to interfere with the light that FIG. 1: Up-Close View of Scales from an Optical Microscope originally reflected off the surface of the film. This inter- ference pattern causes a spectrum of color to be visible from white light. Examples of thin film interference in- clude oil slicks and soap bubbles. Structural coloring oc- curs when the structure of the object itself (with various reflective surfaces) produces an interference resulting in vibrant colors. -

Iridescence in Cooked Venison – an Optical Phenomenon
Journal of Nutritional Health & Food Engineering Research Article Open Access Iridescence in cooked venison – an optical phenomenon Abstract Volume 8 Issue 2 - 2018 Iridescence in single myofibers from roast venison resembled multilayer interference in having multiple spectral peaks that were easily visible under water. The relationship HJ Swatland of iridescence to light scattering in roast venison was explored using the weighted- University of Guelph, Canada ordinate method of colorimetry. In iridescent myofibers, a reflectance ratio (400/700 nm) showing wavelength-dependent light scattering was correlated with HJ Swatland, Designation Professor CIE (Commission International de l’Éclairage) Y%, a measure of overall paleness Correspondence: Emeritus, University of Guelph, 33 Robinson Ave, Guelph, (r=0.48, P< 0.01). Hence, meat iridescence is an optical phenomenon. The underlying Ontario N1H 2Y8, Canada, Tel 519-821-7513, mechanism, subsurface multilayer interference, may be important for meat colorimetry. Email [email protected] venison, iridescence, interference, reflectance, meat color Keywords: Received: August 23, 2017 | Published: March 14, 2018 Introduction balanced pixel hues that have tricked your eyes to appear white; and when we perceive interference colors, complex interference spectra Iridescence is an enigmatic aspect of meat color with some practical trick our eyes again. As the order of interference increases, the colors 1–4 importance for consumers concerned about green colors in meat. appear to change from metallic -

Host-Parasite Interaction of Atlantic Salmon (Salmo Salar) and the Ectoparasite Neoparamoeba Perurans in Amoebic Gill Disease
ORIGINAL RESEARCH published: 31 May 2021 doi: 10.3389/fimmu.2021.672700 Host-Parasite Interaction of Atlantic salmon (Salmo salar) and the Ectoparasite Neoparamoeba perurans in Amoebic Gill Disease † Natasha A. Botwright 1*, Amin R. Mohamed 1 , Joel Slinger 2, Paula C. Lima 1 and James W. Wynne 3 1 Livestock and Aquaculture, CSIRO Agriculture and Food, St Lucia, QLD, Australia, 2 Livestock and Aquaculture, CSIRO Agriculture and Food, Woorim, QLD, Australia, 3 Livestock and Aquaculture, CSIRO Agriculture and Food, Hobart, TAS, Australia Marine farmed Atlantic salmon (Salmo salar) are susceptible to recurrent amoebic gill disease Edited by: (AGD) caused by the ectoparasite Neoparamoeba perurans over the growout production Samuel A. M. Martin, University of Aberdeen, cycle. The parasite elicits a highly localized response within the gill epithelium resulting in United Kingdom multifocal mucoid patches at the site of parasite attachment. This host-parasite response Reviewed by: drives a complex immune reaction, which remains poorly understood. To generate a model Diego Robledo, for host-parasite interaction during pathogenesis of AGD in Atlantic salmon the local (gill) and University of Edinburgh, United Kingdom systemic transcriptomic response in the host, and the parasite during AGD pathogenesis was Maria K. Dahle, explored. A dual RNA-seq approach together with differential gene expression and system- Norwegian Veterinary Institute (NVI), Norway wide statistical analyses of gene and transcription factor networks was employed. A multi- *Correspondence: tissue transcriptomic data set was generated from the gill (including both lesioned and non- Natasha A. Botwright lesioned tissue), head kidney and spleen tissues naïve and AGD-affected Atlantic salmon [email protected] sourced from an in vivo AGD challenge trial. -

Octopus Consciousness: the Role of Perceptual Richness
Review Octopus Consciousness: The Role of Perceptual Richness Jennifer Mather Department of Psychology, University of Lethbridge, Lethbridge, AB T1K 3M4, Canada; [email protected] Abstract: It is always difficult to even advance possible dimensions of consciousness, but Birch et al., 2020 have suggested four possible dimensions and this review discusses the first, perceptual richness, with relation to octopuses. They advance acuity, bandwidth, and categorization power as possible components. It is first necessary to realize that sensory richness does not automatically lead to perceptual richness and this capacity may not be accessed by consciousness. Octopuses do not discriminate light wavelength frequency (color) but rather its plane of polarization, a dimension that we do not understand. Their eyes are laterally placed on the head, leading to monocular vision and head movements that give a sequential rather than simultaneous view of items, possibly consciously planned. Details of control of the rich sensorimotor system of the arms, with 3/5 of the neurons of the nervous system, may normally not be accessed to the brain and thus to consciousness. The chromatophore-based skin appearance system is likely open loop, and not available to the octopus’ vision. Conversely, in a laboratory situation that is not ecologically valid for the octopus, learning about shapes and extents of visual figures was extensive and flexible, likely consciously planned. Similarly, octopuses’ local place in and navigation around space can be guided by light polarization plane and visual landmark location and is learned and monitored. The complex array of chemical cues delivered by water and on surfaces does not fit neatly into the components above and has barely been tested but might easily be described as perceptually rich. -
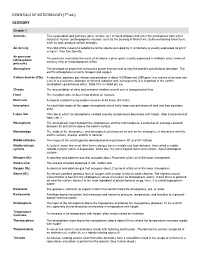
ESSENTIALS of METEOROLOGY (7Th Ed.) GLOSSARY
ESSENTIALS OF METEOROLOGY (7th ed.) GLOSSARY Chapter 1 Aerosols Tiny suspended solid particles (dust, smoke, etc.) or liquid droplets that enter the atmosphere from either natural or human (anthropogenic) sources, such as the burning of fossil fuels. Sulfur-containing fossil fuels, such as coal, produce sulfate aerosols. Air density The ratio of the mass of a substance to the volume occupied by it. Air density is usually expressed as g/cm3 or kg/m3. Also See Density. Air pressure The pressure exerted by the mass of air above a given point, usually expressed in millibars (mb), inches of (atmospheric mercury (Hg) or in hectopascals (hPa). pressure) Atmosphere The envelope of gases that surround a planet and are held to it by the planet's gravitational attraction. The earth's atmosphere is mainly nitrogen and oxygen. Carbon dioxide (CO2) A colorless, odorless gas whose concentration is about 0.039 percent (390 ppm) in a volume of air near sea level. It is a selective absorber of infrared radiation and, consequently, it is important in the earth's atmospheric greenhouse effect. Solid CO2 is called dry ice. Climate The accumulation of daily and seasonal weather events over a long period of time. Front The transition zone between two distinct air masses. Hurricane A tropical cyclone having winds in excess of 64 knots (74 mi/hr). Ionosphere An electrified region of the upper atmosphere where fairly large concentrations of ions and free electrons exist. Lapse rate The rate at which an atmospheric variable (usually temperature) decreases with height. (See Environmental lapse rate.) Mesosphere The atmospheric layer between the stratosphere and the thermosphere. -

Tenacibaculum Maritimum, Causal Agent of Tenacibaculosis in Marine Fish
# 70 JANUARY 2019 Tenacibaculum maritimum, causal agent of tenacibaculosis in marine fish ICES IDENTIFICATION LEAFLETS FOR DISEASES AND PARASITES IN FISH AND SHELLFISH ICES INTERNATIONAL COUNCIL FOR THE EXPLORATION OF THE SEA CIEM CONSEIL INTERNATIONAL POUR L’EXPLORATION DE LA MER ICES IDENTIFICATION LEAFLETS FOR DISEASES AND PARASITES OF FISH AND SHELLFISH NO. 70 JANUARY 2019 Tenacibaculum maritimum, causal agent of tenacibaculosis in marine fish Original by Y. Santos, F. Pazos and J. L. Barja (No. 55) Revised by Simon R. M. Jones and Lone Madsen International Council for the Exploration of the Sea Conseil International pour l’Exploration de la Mer H. C. Andersens Boulevard 44–46 DK-1553 Copenhagen V Denmark Telephone (+45) 33 38 67 00 Telefax (+45) 33 93 42 15 www.ices.dk [email protected] Recommended format for purposes of citation: ICES 2019. Tenacibaculum maritimum, causal agent of tenacibaculosis in marine fish. Orig- inal by Santos Y., F. Pazos and J. L. Barja (No. 55), Revised by Simon R. M. Jones and Lone Madsen. ICES Identification Leaflets for Diseases and Parasites of Fish and Shell- fish. No. 70. 5 pp. http://doi.org/10.17895/ices.pub.4681 Series Editor: Neil Ruane. Prepared under the auspices of the ICES Working Group on Pathology and Diseases of Marine Organisms. The material in this report may be reused for non-commercial purposes using the rec- ommended citation. ICES may only grant usage rights of information, data, images, graphs, etc. of which it has ownership. For other third-party material cited in this re- port, you must contact the original copyright holder for permission. -

First Isolation of Virulent Tenacibaculum Maritimum Strains
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.15.435441; this version posted March 15, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 First isolation of virulent Tenacibaculum maritimum 2 strains from diseased orbicular batfish (Platax orbicularis) 3 farmed in Tahiti Island 4 Pierre Lopez 1¶, Denis Saulnier 1¶*, Shital Swarup-Gaucher 2, Rarahu David 2, Christophe Lau 2, 5 Revahere Taputuarai 2, Corinne Belliard 1, Caline Basset 1, Victor Labrune 1, Arnaud Marie 3, Jean 6 François Bernardet 4, Eric Duchaud 4 7 8 9 1 Ifremer, IRD, Institut Louis‐Malardé, Univ Polynésie française, EIO, Labex Corail, F‐98719 10 Taravao, Tahiti, Polynésie française, France 11 2 DRM, Direction des ressources marines, Fare Ute Immeuble Le caill, BP 20 – 98713 Papeete, Tahiti, 12 Polynésie française 13 3 Labofarm Finalab Veterinary Laboratory Group, 4 rue Théodore Botrel, 22600 Loudéac, France 14 4 Unité VIM, INRAE, Université Paris-Saclay, 78350 Jouy-en-Josas, France 15 * Corresponding author 16 E-mail: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.03.15.435441; this version posted March 15, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 17 Abstract 18 The orbicular batfish (Platax orbicularis), also called 'Paraha peue' in Tahitian, is the most important 19 marine fish species reared in French Polynesia. -

Full Article in Pdf Format
DISEASES OF AQUATIC ORGANISMS Vol. 58: 1–8, 2004 Published January 28 Dis Aquat Org Phenotyphic characterization and description of two major O-serotypes in Tenacibaculum maritimum strains from marine fishes Ruben Avendaño-Herrera, Beatriz Magariños, Sonia López-Romalde, Jesús L. Romalde, Alicia E. Toranzo* Departamento de Microbiología y Parasitología, Facultad de Biología, Universidad de Santiago, 15782 Santiago de Compostela, Spain ABSTRACT: Tenacibaculum maritimum is the etiological agent of marine flexibacteriosis disease, with the potential to cause severe mortalities in various cultured marine fishes. The development of effective preventive measures (i.e. vaccination) requires biochemical, serological and genetic knowl- edge of the pathogen. With this aim, the biochemical and antigenic characteristics of T. maritimum strains isolated from sole, turbot and gilthead sea bream were analysed. Rabbit antisera were pre- pared against sole and turbot strains to examine the antigenic relationships between the 29 isolates and 3 reference strains. The results of the slide agglutination test, dot-blot assay and immunoblotting of lipopolysaccharides (LPS) and membrane proteins were evaluated. All bacteria studied were biochemically identical to the T. maritimum reference strains. The slide agglutination assays using O-antigens revealed cross-reaction for all strains regardless of the host species and serum employed. However, when the dot-blot assays were performed, the existence of antigenic heterogeneity was demonstrated. This heterogeneity was supported by immunoblot analysis of the LPS, which clearly revealed 2 major serological groups that were distinguishable without the use of absorbed antiserum: Serotypes O1 and O2. These 2 serotypes seem to be host-specfic. In addition, 2 sole isolates and the Japanese reference strains displayed cross-reaction with both sera in all serological assays, and are considered to constitute a minor serotype, O1/O2. -

Rainbow Peacock Spiders Inspire Miniature Super-Iridescent Optics
ARTICLE DOI: 10.1038/s41467-017-02451-x OPEN Rainbow peacock spiders inspire miniature super- iridescent optics Bor-Kai Hsiung 1,8, Radwanul Hasan Siddique 2, Doekele G. Stavenga 3, Jürgen C. Otto4, Michael C. Allen5, Ying Liu6, Yong-Feng Lu 6, Dimitri D. Deheyn 5, Matthew D. Shawkey 1,7 & Todd A. Blackledge1 Colour produced by wavelength-dependent light scattering is a key component of visual communication in nature and acts particularly strongly in visual signalling by structurally- 1234567890 coloured animals during courtship. Two miniature peacock spiders (Maratus robinsoni and M. chrysomelas) court females using tiny structured scales (~ 40 × 10 μm2) that reflect the full visual spectrum. Using TEM and optical modelling, we show that the spiders’ scales have 2D nanogratings on microscale 3D convex surfaces with at least twice the resolving power of a conventional 2D diffraction grating of the same period. Whereas the long optical path lengths required for light-dispersive components to resolve individual wavelengths constrain current spectrometers to bulky sizes, our nano-3D printed prototypes demonstrate that the design principle of the peacock spiders’ scales could inspire novel, miniature light-dispersive components. 1 Department of Biology and Integrated Bioscience Program, The University of Akron, Akron, OH 44325, USA. 2 Department of Medical Engineering, California Institute of Technology, Pasadena, CA 91125, USA. 3 Department of Computational Physics, University of Groningen, 9747 AG Groningen, The Netherlands. 4 19 Grevillea Avenue, St. Ives, NSW 2075, Australia. 5 Scripps Institution of Oceanography (SIO), University of California, San Diego, La Jolla, CA 92093, USA. 6 Department of Electrical and Computer Engineering, University of Nebraska-Lincoln, Lincoln, NE 68588, USA. -

Penaeus Monodon
www.nature.com/scientificreports OPEN Bacterial analysis in the early developmental stages of the black tiger shrimp (Penaeus monodon) Pacharaporn Angthong1,3, Tanaporn Uengwetwanit1,3, Sopacha Arayamethakorn1, Panomkorn Chaitongsakul2, Nitsara Karoonuthaisiri1 & Wanilada Rungrassamee1* Microbial colonization is an essential process in the early life of animal hosts—a crucial phase that could help infuence and determine their health status at the later stages. The establishment of bacterial community in a host has been comprehensively studied in many animal models; however, knowledge on bacterial community associated with the early life stages of Penaeus monodon (the black tiger shrimp) is still limited. Here, we examined the bacterial community structures in four life stages (nauplius, zoea, mysis and postlarva) of two black tiger shrimp families using 16S rRNA amplicon sequencing by a next-generation sequencing. Although the bacterial profles exhibited diferent patterns in each developmental stage, Bacteroidetes, Proteobacteria, Actinobacteria and Planctomycetes were identifed as common bacterial phyla associated with shrimp. Interestingly, the bacterial diversity became relatively stable once shrimp developed to postlarvae (5-day-old and 15-day- old postlarval stages), suggesting an establishment of the bacterial community in matured shrimp. To our knowledge, this is the frst report on bacteria establishment and assembly in early developmental stages of P. monodon. Our fndings showed that the bacterial compositions could be shaped by diferent host developmental stages where the interplay of various host-associated factors, such as physiology, immune status and required diets, could have a strong infuence. Te shrimp aquaculture industry is one of the key sectors to supply food source to the world’s growing pop- ulation. -

Iridescent Color: from Nature to the Painter's Palette
Downloaded from http://www.mitpressjournals.org/doi/pdf/10.1162/LEON_a_00114 by guest on 25 September 2021 Technical ar T i c l e ce, Tech N e Iridescent Color: From Nature I , Sc to the Painter’s Palette T a b s T r a c T o: Ar nan The shifting rainbow hues of Franziska Schenk iridescence have, until recently, remained exclusive to nature. and Andrew Parker Now, the latest advances in nanotechnology enable the introduction of novel, bio- inspired color-shifting flakes into painting—thereby affording artists potential access to the full spectacle of iridescence. Unfortunately, existing rules rtists have never captured a color as dazzling selves of two distinct types: those of easel painting do not apply A that are stable attributes of material to the new medium; but, as and dynamic as the metallic blue of the Morpho butterfly, which nature inspired the technol- is visible for up to a quarter of a mile. Now, with rapid ad- substances, and those that are “acci- dental,” such as the evanescent col- ogy, an exploration of natural vances in nanoscience and technology, we are beginning to ors of the rainbow and the colors of phenomena can best inform unravel nature’s ingenious manipulation of the flow of light. some birds’ feathers, which change how to overcome this hurdle. Scientific research into natural nanoscale architectures, ca- according to the viewpoint of the Thus, by adopting a biomimetic pable of producing eye-catching optical effects, has led to the spectator [2]. approach, this paper outlines the optical principles underly- development of an ever-expanding range of comparable syn- ing iridescence and provides thetic structures. -

Tenacibaculum Adriaticum Sp. Nov., from a Bryozoan in the Adriatic Sea
International Journal of Systematic and Evolutionary Microbiology (2008), 58, 542–547 DOI 10.1099/ijs.0.65383-0 Tenacibaculum adriaticum sp. nov., from a bryozoan in the Adriatic Sea Herwig Heindl, Jutta Wiese and Johannes F. Imhoff Correspondence Kieler Wirkstoff-Zentrum (KiWiZ) at the Leibniz-Institute for Marine Sciences, Am Kiel-Kanal 44, Johannes F. Imhoff D-24106 Kiel, Germany [email protected] A rod-shaped, translucent yellow-pigmented, Gram-negative bacterium, strain B390T, was isolated from the bryozoan Schizobrachiella sanguinea collected in the Adriatic Sea, near Rovinj, Croatia. 16S rRNA gene sequence analysis indicated affiliation to the genus Tenacibaculum, with sequence similarity levels of 94.8–97.3 % to type strains of species with validly published names. It grew at 5–34 6C, with optimal growth at 18–26 6C, and only in the presence of NaCl or sea salts. In contrast to other type strains of the genus, strain B390T was able to hydrolyse aesculin. The predominant menaquinone was MK-6 and major fatty acids were iso-C15 : 0, iso-C15 : 0 3-OH and iso-C15 : 1. The DNA G+C content was 31.6 mol%. DNA–DNA hybridization and comparative physiological tests were performed with type strains Tenacibaculum aestuarii JCM 13491T and Tenacibaculum lutimaris DSM 16505T, since they exhibit 16S rRNA gene sequence similarities above 97 %. These data, as well as phylogenetic analyses, suggest that strain B390T (5DSM 18961T 5JCM 14633T) should be classified as the type strain of a novel species within the genus Tenacibaculum, for which the name Tenacibaculum adriaticum sp. nov. is proposed.