Nucleoside Modifications Suppress RNA Activation of Cytoplasmic RNA Sensors
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Split Deoxyribozyme Probe for Efficient Detection of Highly Structured RNA Targets
University of Central Florida STARS Honors Undergraduate Theses UCF Theses and Dissertations 2018 Split Deoxyribozyme Probe For Efficient Detection of Highly Structured RNA Targets Sheila Raquel Solarez University of Central Florida Part of the Biochemistry Commons, and the Biology Commons Find similar works at: https://stars.library.ucf.edu/honorstheses University of Central Florida Libraries http://library.ucf.edu This Open Access is brought to you for free and open access by the UCF Theses and Dissertations at STARS. It has been accepted for inclusion in Honors Undergraduate Theses by an authorized administrator of STARS. For more information, please contact [email protected]. Recommended Citation Solarez, Sheila Raquel, "Split Deoxyribozyme Probe For Efficient Detection of Highly Structured RNA Targets" (2018). Honors Undergraduate Theses. 311. https://stars.library.ucf.edu/honorstheses/311 SPLIT DEOXYRIBOZYME PROBE FOR EFFICIENT DETECTION OF HIGHLY STRUCTURED RNA TARGETS By SHEILA SOLAREZ A thesis submitted in partial fulfillment of the requirements for the Honors in the Major Program in Biological Sciences in the College of Sciences and the Burnett Honors College at the University of Central Florida Orlando, Florida Spring Term, 2018 Thesis Chair: Yulia Gerasimova, PhD ABSTRACT Transfer RNAs (tRNAs) are known for their role as adaptors during translation of the genetic information and as regulators for gene expression; uncharged tRNAs regulate global gene expression in response to changes in amino acid pools in the cell. Aminoacylated tRNAs play a role in non-ribosomal peptide bond formation, post-translational protein labeling, modification of phospholipids in the cell membrane, and antibiotic biosynthesis. [1] tRNAs have a highly stable structure that can present a challenge for their detection using conventional techniques. -

Mrna Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection
International Journal of Molecular Sciences Review mRNA Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection 1, 1, 2 1,3, Shuqin Xu y, Kunpeng Yang y, Rose Li and Lu Zhang * 1 State Key Laboratory of Genetic Engineering, Institute of Genetics, School of Life Science, Fudan University, Shanghai 200438, China; [email protected] (S.X.); [email protected] (K.Y.) 2 M.B.B.S., School of Basic Medical Sciences, Peking University Health Science Center, Beijing 100191, China; [email protected] 3 Shanghai Engineering Research Center of Industrial Microorganisms, Shanghai 200438, China * Correspondence: [email protected]; Tel.: +86-13524278762 These authors contributed equally to this work. y Received: 30 July 2020; Accepted: 30 August 2020; Published: 9 September 2020 Abstract: Messenger ribonucleic acid (mRNA)-based drugs, notably mRNA vaccines, have been widely proven as a promising treatment strategy in immune therapeutics. The extraordinary advantages associated with mRNA vaccines, including their high efficacy, a relatively low severity of side effects, and low attainment costs, have enabled them to become prevalent in pre-clinical and clinical trials against various infectious diseases and cancers. Recent technological advancements have alleviated some issues that hinder mRNA vaccine development, such as low efficiency that exist in both gene translation and in vivo deliveries. mRNA immunogenicity can also be greatly adjusted as a result of upgraded technologies. In this review, we have summarized details regarding the optimization of mRNA vaccines, and the underlying biological mechanisms of this form of vaccines. Applications of mRNA vaccines in some infectious diseases and cancers are introduced. It also includes our prospections for mRNA vaccine applications in diseases caused by bacterial pathogens, such as tuberculosis. -

Investigation of Overhauser Effects Between Pseudouridine and Water Protons in RNA Helices
Investigation of Overhauser effects between pseudouridine and water protons in RNA helices Meredith I. Newby* and Nancy L. Greenbaum*†‡ *Department of Chemistry and Biochemistry and †Institute of Molecular Biophysics, Florida State University, Tallahassee, FL 32306-4390 Communicated by Michael Kasha, Florida State University, Tallahassee, FL, August 8, 2002 (received for review July 2, 2002) The inherent chemical properties of RNA molecules are expanded by posttranscriptional modification of specific nucleotides. Pseudouridine (), the most abundant of the modified bases, features an additional imino group, NH1, as compared with uri- dine. When forms a Watson–Crick base pair with adenine in an RNA helix, NH1 is positioned within the major groove. The pres- ence of often increases thermal stability of the helix or loop in which it is found [Hall, K. B. & McLaughlin, L. (1992) Nucleic Acids Res. 20, 1883–1889]. X-ray crystal structures of transfer RNAs [e.g., Arnez, J. & Steitz, T. (1994) Biochemistry 33, 7560–7567] have depicted water molecules bridging NH1 groups and nearby phos- phate oxygen atoms, but direct evidence for this interaction in solution has not been acquired. Toward this end, we have used a rotating-frame Overhauser effect spectroscopy-type NMR pulse sequence with a CLEAN chemical-exchange spectroscopy spin-lock pulse train [Hwang, T.-L., Mori, S., Shaka, A. J. & van Zijl, P. C. M. Fig. 1. Schematic structures of U and bases. R, ribose. The base is a uracil (1997) J. Am. Chem. Soc. 119, 6203–6204] to test for NH1–water rotated about the 3–6 ring axis, so that is has a COC base–sugar linkage and cross-relaxation effects within two RNA helices: (i) a complemen- an additional protonated ring nitrogen. -

Abou Alezz, Monah Characterization of the Genomic and Splicing Features of Long 51 Non-Coding Rnas Using Bioinformatics Approaches
Posters A-Z Abou Alezz, Monah Characterization of the genomic and splicing features of long 51 non-coding RNAs using bioinformatics approaches Aguilera-Cortés, Paulina Identification of cellular lncRNAs associated to the HIV-1 genomic RNA 52 by interactome capture Ahram, Mamoun The androgen, dihydrotestosterone, induces chemoresistance of triple 53 negative breast cancer cells independent of ABCG2 and microRNA-328-3p Ahunbay, Esra Watching the self-catalyzed splicing of a group II intron with 54 single-molecule sensitivity Alemu, Endalkachew Integrated Global Mapping of miRNA:target-RNA Interactions 55 Ali, Tamer The lncRNA locus Handsdown regulates cardiac gene programs and is 56 essential for early mouse development Alzahrani, Salma Revisiting the sRNAome Under a Revised MicroRNA Annotation Criteria 57 Amorim, Marcella Dissecting the threshold-based readout of mobile small RNA gradients 58 Arnoldi, Michele SINEUPs technology: a new route to possibly treat 59 haploinsufficiency-induced Epilepsy and Autism Spectrum Disorders (ASDs) Azhikina, Tatyana Small RNA F6 influences Mycobacterium smegmatis entry into 60 dormancy EMBO | EMBL Symposium: The Non-Coding Genome Barasa, Sheila Olendo Long non-coding RNAs in High Grade Serous Ovarian Carcinoma 61 Behera, Alok LIN28 as a new drug target 62 Behrmann, Iris Modulation of the IL-6-Signaling Pathway in Liver Cells by miRNAs 63 Targeting gp130, JAK1, and/or STAT3 Bencurova, Petra Inhibition of miRNA-129-2-3p increases risk and severity of seizures in 64 developing brain Bica, Cecilia The emerging -

Expanding the Genetic Code Lei Wang and Peter G
Reviews P. G. Schultz and L. Wang Protein Science Expanding the Genetic Code Lei Wang and Peter G. Schultz* Keywords: amino acids · genetic code · protein chemistry Angewandte Chemie 34 2005 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim DOI: 10.1002/anie.200460627 Angew. Chem. Int. Ed. 2005, 44,34–66 Angewandte Protein Science Chemie Although chemists can synthesize virtually any small organic molecule, our From the Contents ability to rationally manipulate the structures of proteins is quite limited, despite their involvement in virtually every life process. For most proteins, 1. Introduction 35 modifications are largely restricted to substitutions among the common 20 2. Chemical Approaches 35 amino acids. Herein we describe recent advances that make it possible to add new building blocks to the genetic codes of both prokaryotic and 3. In Vitro Biosynthetic eukaryotic organisms. Over 30 novel amino acids have been genetically Approaches to Protein encoded in response to unique triplet and quadruplet codons including Mutagenesis 39 fluorescent, photoreactive, and redox-active amino acids, glycosylated 4. In Vivo Protein amino acids, and amino acids with keto, azido, acetylenic, and heavy-atom- Mutagenesis 43 containing side chains. By removing the limitations imposed by the existing 20 amino acid code, it should be possible to generate proteins and perhaps 5. An Expanded Code 46 entire organisms with new or enhanced properties. 6. Outlook 61 1. Introduction The genetic codes of all known organisms specify the same functional roles to amino acid residues in proteins. Selectivity 20 amino acid building blocks. These building blocks contain a depends on the number and reactivity (dependent on both limited number of functional groups including carboxylic steric and electronic factors) of a particular amino acid side acids and amides, a thiol and thiol ether, alcohols, basic chain. -
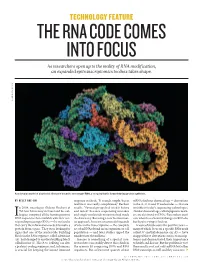
THE RNA CODE COMES INTO FOCUS As Researchers Open up to the Reality of RNA Modification, an Expanded Epitranscriptomics Toolbox Takes Shape
TECHNOLOGY FEATURE THE RNA CODE COMES INTO FOCUS As researchers open up to the reality of RNA modification, an expanded epitranscriptomics toolbox takes shape. LAGUNA DESIGN/SPL LAGUNA A molecular model of a bacterial ribosome bound to messenger RNA, a complex that is formed during protein synthesis. BY KELLY RAE CHI response in check. “It sounds simple, but in mRNAs harbour chemical tags — decorations real life it was really complicated,” Rechavi to the A, C, G and U nucleotides — that are n 2004, oncologist Gideon Rechavi at recalls. “Several groups had tried it before invisible to today’s sequencing technologies. Tel Aviv University in Israel and his col- and failed” because sequencing mistakes (Similar chemical tags, called epigenetic mark- leagues compared all the human genomic and single-nucleotide mutations had made ers, are also found on DNA.) Researchers aren’t IDNA sequences then available with their cor- the data noisy. But using a new bioinformat- sure what these chemical changes in RNA do, responding messenger RNAs — the molecules ics approach, his team uncovered thousands but they’re trying to find out. that carry the information needed to make a of sites in the transcriptome — the complete A wave of studies over the past five years — protein from a gene. They were looking for set of mRNAs found in an organism or cell many of which focus on a specific RNA mark signs that one of the nucleotide building population — and later studies upped the called N6-methyladenosine (m6A) — have blocks in the RNA sequence, called adenosine number into the millions1. -

The Human Ortholog of Archaeal Pus10 Produces Pseudouridine 54 in Select Trnas Where Its Recognition Sequence Contains a Modified Residue
Downloaded from rnajournal.cshlp.org on October 7, 2021 - Published by Cold Spring Harbor Laboratory Press The human ortholog of archaeal Pus10 produces pseudouridine 54 in select tRNAs where its recognition sequence contains a modified residue MANISHA DEOGHARIA,1 SHAONI MUKHOPADHYAY, ARCHI JOARDAR,2 and RAMESH GUPTA Department of Biochemistry and Molecular Biology, Southern Illinois University, Carbondale, Illinois 62901-4413, USA ABSTRACT The nearly conserved U54 of tRNA is mostly converted to a version of ribothymidine (T) in Bacteria and eukaryotes and to a version of pseudouridine (Ψ) in Archaea. Conserved U55 is nearly always modified to Ψ55 in all organisms. Orthologs of TrmA and TruB that produce T54 and Ψ55, respectively, in Bacteria and eukaryotes are absent in Archaea. Pus10 produces both Ψ54 and Ψ55 in Archaea. Pus10 orthologs are found in nearly all sequenced archaeal and most eukaryal genomes, but not in yeast and bacteria. This coincides with the presence of Ψ54 in most archaeal tRNAs and some animal tRNAs, but its absence from yeast and bacteria. Moreover, Ψ54 is found in several tRNAs that function as primers for retroviral DNA syn- thesis. Previously, no eukaryotic tRNA Ψ54 synthase had been identified. We show here that human Pus10 can produce Ψ54 in select tRNAs, including tRNALys3, the primer for HIV reverse transcriptase. This synthase activity of Pus10 is restrict- ed to the cytoplasm and is distinct from nuclear Pus10, which is known to be involved in apoptosis. The sequence GUUCAm1AAUC (m1A is 1-methyladenosine) at position 53–61 of tRNA along with a stable acceptor stem results in max- imum Ψ54 synthase activity. -

Pseudouridine Synthases Modify Human Pre-Mrna Co-Transcriptionally and Affect Splicing
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.29.273565; this version posted August 31, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Pseudouridine synthases modify human pre-mRNA co-transcriptionally and affect splicing Authors: Nicole M. Martinez1, Amanda Su1, Julia K. Nussbacher2,3,4, Margaret C. Burns2,3,4, Cassandra Schaening5, Shashank Sathe2,3,4, Gene W. Yeo2,3,4* and Wendy V. Gilbert1* Authors and order TBD with final revision. Affiliations: 1Yale School of Medicine, Department of Molecular Biophysics & Biochemistry, New Haven, CT 06520, USA. 2Department of Cellular and Molecular Medicine, University of California, San Diego, La Jolla, CA 92037, USA. 3Stem Cell Program, University of California, San Diego, La Jolla, CA 92037, USA. 4Institute for Genomic Medicine, University of California, San Diego, La Jolla, CA 92037, USA. 5Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02142, USA. *Correspondence to: [email protected], [email protected] Abstract: Eukaryotic messenger RNAs are extensively decorated with modified nucleotides and the resulting epitranscriptome plays important regulatory roles in cells 1. Pseudouridine (Ψ) is a modified nucleotide that is prevalent in human mRNAs and can be dynamically regulated 2–5. However, it is unclear when in their life cycle RNAs become pseudouridylated and what the endogenous functions of mRNA pseudouridylation are. To determine if pseudouridine is added co-transcriptionally, we conducted pseudouridine profiling 2 on chromatin-associated RNA to reveal thousands of intronic pseudouridines in nascent pre-mRNA at locations that are significantly associated with alternatively spliced exons, enriched near splice sites, and overlap hundreds of binding sites for regulatory RNA binding proteins. -

Mrna, Rrna and Trna Types of RNA: Mrna, Rrna and Trna
Types of RNA: mRNA, rRNA and tRNA Types of RNA: mRNA, rRNA and tRNA By Susha Cheriyedath, M.Sc. Reviewed by Michael Greenwood, M.Sc. RNA or ribonucleic acid is a polymer of nucleotides that is made up of a ribose sugar, a phosphate, and bases such as adenine, guanine, cytosine, and uracil. It plays a crucial role in gene expression by acting as the intermediate between the genetic information encoded by DNA and proteins. Designua | Shutterstock RNA has a structure very similar to that of DNA. The key difference in RNA structure is that the ribose sugar in RNA possesses a hydroxyl (OH) group that is absent in DNA. Types of RNA In both prokaryotes and eukaryotes, there are three main types of RNA – messenger RNA (mRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA). These 3 types of RNA are discussed below. P Saved from URL: https://www.news-medical.net/life-sciences/-Types-of-RNA-mRNA-rRNA-and-tRNA.aspx 1/5 Types of RNA: mRNA, rRNA and tRNA Messenger RNA (mRNA) mRNA accounts for just 5% of the total RNA in the cell. mRNA is the most heterogeneous of the 3 types of RNA in terms of both base sequence and size. It carries complimentary genetic code copied, from DNA during transcription, in the form of triplets of nucleotides called codons. Each codon specifies a particular amino acid, though one amino acid may be coded for by many different codons. Although there are 64 possible codons or triplet bases in the genetic code, only 20 of them represent amino acids. -

Transcription Study Guide This Study Guide Is a Written Version of the Material You Have Seen Presented in the Transcription Unit
Transcription Study Guide This study guide is a written version of the material you have seen presented in the transcription unit. The cell’s DNA contains the instructions for carrying out the work of the cell. These instructions are used by the cell’s protein-making machinery to create proteins. If the cell’s DNA were directly read by the protein-making machinery, however, it could be damaged and the process would be slow and cumbersome. The cell avoids this problem by copying genetic information from its DNA into an intermediate called messenger RNA (mRNA). It is this mRNA that is read by the cell’s protein-making machinery. This process is called transcription. Components In this section you will be introduced to the components involved in the process of RNA synthesis, called transcription. This process requires an enzyme that uses many nucleotide bases to copy the instructions present in DNA into an intermediate messenger RNA molecule. RNA What is RNA? · Like DNA, RNA is a polymer made up of nucleotides. · Unlike DNA, which is composed of two strands of nucleotides twisted together, RNA is single-stranded. It can also sometimes fold into complex three-dimensional structures. · RNA contains the same nucleotides as DNA, with the substitution of uraciluridine (U) for thymidine (T). · RNA is chemically different from DNA so that the cell can easily tell the two apart. · In this animation, you will see one type of RNA, messenger RNA, being put together. · There are three types of RNA: mRNA, which you will read more about; tRNA, which is used in the translation process, and rRNA, which acts as a structural element in the ribosome (a translation component). -

Isolation of RNA with Properties of Messenger RNA from Cerebral Polyribosomes* Claire E
Proceedings of the National Academy of Sciences Vol. 67, No. 2, pp. 644-651, October 1970 Isolation of RNA with Properties of Messenger RNA from Cerebral Polyribosomes* Claire E. Zomzelyt, Sidney Roberts, and Susan Peache DEPARTMENT OF BIOLOGICAL CHEMISTRY, SCHOOL OF MEDICINE, AND THE BRAIN RESEARCH INSTI- TUTE, UNIVERSITY OF CALIFORNIA CENTER FOR THE HEALTH SCIENCES, LOS ANGELES, CALIFORNIA 90024 Communicated by H. W. Magoun, July 24, 1970 Abstract. RNA which dissociated from purified cerebral polyribosomes of adult rats in the presence of EDTA was isolated by fractionation in a dis- continuous sucrose gradient. The yield was 2% of the total polyribosomal RNA. The base composition resembled the complementary values for rat DNA and was very different from base compositions of ribosomal RNA and transfer RNA. This RNA fraction contained a large proportion of molecules which were rapidly labeled in vivo and hybridized to homologous DNA. The polyribosomal RNA preparation also exhibited high template activity in a cerebral cell-free system which had previously been stripped of the capacity to incorporate amino acids in the absence of added messenger RNA (mRNA). Sedimentation analysis revealed only two peaks, with coefficients of approximately 8 S and 16 S. The data indicate that RNA with the properties of mRNA can be selectively isolated from cerebral polyribosomes under mild conditions which avoid degradation. Numerous studies support the concept that alterations in RNA and protein synthesis are associated with information transfer and storage in the central nervous system. Activation of the synthesis of specific proteins is suggested by the reports that new RNA with altered base composition, presumably mRNA, is formed during the acquisition phase of learning. -

A Dissertation Entitled Ribonucleic Acids in Disease Etiology and Drug Discovery by Immaculate Sappy Submitted to the Graduate F
A Dissertation entitled Ribonucleic Acids in Disease Etiology and Drug Discovery by Immaculate Sappy Submitted to the Graduate Faculty as partial fulfillment of the requirements for the Doctor of Philosophy Degree in Medicinal Chemistry ________________________________________ Amanda C. Bryant-Friedrich, Ph.D., Committee Chair ________________________________________ Zahoor A Shah, Ph.D., Committee Member ________________________________________ Steven M Peseckis, Ph.D., Committee Member ________________________________________ Caren Steinmiller, Ph.D., Committee Member ________________________________________ Amanda Bryant-Friedrich, PhD, Dean College of Graduate Studies The University of Toledo December 2019 Copyright 2019, Immaculate Sappy This document is copyrighted material. Under copyright law, no parts of this document may be reproduced without the expressed permission of the author. An Abstract of Ribonucleic Acids in Disease Etiology and Drug Discovery by Immaculate Sappy Submitted to the Graduate Faculty as partial fulfillment of the requirements for the Doctor of Philosophy Degree in Medicinal Chemistry The University of Toledo December 2019 Pseudouridine (Ψ), the 5-ribosyl isomer of uridine (U) is the most abundant nucleic acid modification found in all domains of life and all types of RNA. Studies have shown that, urinary levels of pseudouridine are higher in Alzheimer’s Disease (AD) patients and that RNA oxidation is a major component in the pathogenesis of Alzheimer’s Disease (AD) and other neurodegenerative disorders. Therefore, there is a potential correlation between higher urinary levels of pseudouridine in AD patients and oxidative stress. Hence, subjecting pseudouridine to oxidative conditions may provide some key information about the role of this nucleoside in RNA related processes and its role in disease etiology. Besides neurodegenerative disorders, antibiotic resistance is an additional threat to human health.