Genetic Diversity and Geographic Distribution of Bat-Borne Hantaviruses
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cryptic Phylogeographic History Sheds Light on the Generation of Species Diversity in Sky-Island Mountains
bioRxiv preprint doi: https://doi.org/10.1101/199786; this version posted October 7, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Cryptic phylogeographic history sheds light on the generation of species diversity in sky-island mountains Kai He1, 2, 3, #, Tao Wan 1, 4, Klaus-Peter Koepfli 5, 6, Wei Jin7, Shao-Ying Liu7, Xue-Long Jiang1, # 1 State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China 2 Department of Biological Sciences, University of Manitoba, Winnipeg, MN R3T R3V, Canada 3 The Kyoto University Museum, Kyoto University, Kyoto 606-8501, Japan. 4 Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming, Yunnan 650223, China 5 Smithsonian Conservation Biology Institute, National Zoological Park, DC 20008, USA 6 Theodosius Dobzhansky Center for Genome Bioinformatics, Saint Petersburg State University, St. Petersburg 199034, RUSSIA 7 Sichuan Academy of Forest, Chengdu 610081, Sichuan, China Keywords: allopatry, Approximate Bayesian Computation, cryptic corridor, interglacial refugia, niche modeling, species delimitation Running title: Cryptic sky-island phylogeography # Correspondence: Kai He and Xue-Long Jiang, Fax: 86 871 6512 5226; E-mails: [email protected], [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/199786; this version posted October 7, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Morphometrical Variations of Malaysian Hipposideros Species
Malaysian Journal of Mathematical Sciences 6(1): 47-57 (2012) Morphometrical Variations of Malaysian Hipposideros Species Siti Nurlydia Sazali, Charlie J. Laman and M.T. Abdullah Department of Zoology, Faculty of Resource Science and Technology, Universiti Malaysia Sarawak, 94300 Kota Samarahan, Sarawak, Malaysia E-mail: [email protected] ABSTRACT A study on the morphometrical variations among four Malaysian Hipposideros species was conducted using voucher specimens deposited in Universiti Malaysia Sarawak (UNIMAS) Zoological Museum and the Department of Widlife and National Park (DWNP) Kuala Lumpur. Twenty two individuals from four species of Hipposideros ater , H. bicolor , H. cineraceus and H. dyacorum were morphologically measured, in which a total of 27 linear parameters of body, skull and dentals of each were appropriately recorded. The statistical data were later subjected to discriminant function analysis (DFA) and canonical variate analysis (CVA) using SPSS version 15.0 and unweighted pair-group method average (UPGMA) cluster analysis using Minitab version 14.4. The highest character loadings observed in Function l, Function 2 and Function 3 were the forearm length (FA), the third digit second phalanx length (D3P2L) and the palatal length (PL) with standardised canonical discriminant function coefficient values of 21.910, 5.770 and 5.095, respectively. These three characters were identified as the best diagnostic features for discriminating these closely related species of Hipposideros . Hence, this morphometric approach could be a promising tool as an alternative to the molecular DNA analysis for identification of Chiroptera species. Keywords: Hipposideros , morphometric, discriminant function analysis cluster analysis, species identification. 1. INTRODUCTION Bats belong to the order Chiroptera and can be distinguished from all other mammals by their ability to fly, which is a result of the modification of their forelimbs into wings (Payne et al . -

Interglacial Refugia Preserved High Genetic Diversity of the Chinese Mole Shrew in the Mountains of Southwest China
Heredity (2016) 116, 23–32 & 2016 Macmillan Publishers Limited All rights reserved 0018-067X/16 www.nature.com/hdy ORIGINAL ARTICLE Interglacial refugia preserved high genetic diversity of the Chinese mole shrew in the mountains of southwest China KHe1,2, N-Q Hu1,3, X Chen4,5, J-T Li6 and X-L Jiang1 The mountains of southwest China (MSC) harbor extremely high species diversity; however, the mechanism behind this diversity is unknown. We investigated to what degree the topography and climate change shaped the genetic diversity and diversification in these mountains, and we also sought to identify the locations of microrefugia areas in these mountains. For these purposes, we sampled extensively to estimate the intraspecific phylogenetic pattern of the Chinese mole shrew (Anourosorex squamipes)in southwest China throughout its range of distribution. Two mitochondrial genes, namely, cytochrome b (CYT B) and NADH dehydrogenase subunit 2 (ND2), from 383 archived specimens from 43 localities were determined for phylogeographic and demographic analyses. We used the continuous-diffusion phylogeographic model, extensive Bayesian skyline plot species distribution modeling (SDM) and approximate Bayesian computation (ABC) to explore the changes in population size and distribution through time of the species. Two phylogenetic clades were identified, and significantly higher genetic diversity was preserved in the southern subregion of the mountains. The results of the SDM, continuous-diffusion phylogeographic model, extensive Bayesian skyline plot and ABC analyses were congruent and supported that the Last Interglacial Maximum (LIG) was an unfavorable period for the mole shrews because of a high degree of seasonality; A. squamipes survived in isolated interglacial refugia mainly located in the southern subregion during the LIG and rapidly expanded during the last glacial period. -

BANDICOTA INDICA, the BANDICOOT RAT 3.1 The
CHAPTER THREE BANDICOTA INDICA, THE BANDICOOT RAT 3.1 The Living Animal 3.1.1 Zoology Rats and mice (family Muridae) are the most common and well-known rodents, not only of the fi elds, cultivated areas, gardens, and storage places but especially so of the houses. Though there are many genera and species, their general appearance is pretty the same. Rats are on average twice as large as mice (see Chapter 31). The bandicoot is the largest rat on the Indian subcontinent, with a body and head length of 30–40 cm and an equally long tail; this is twice as large as the black rat or common house rat (see section 3.1.2 below). This large size immediately distinguishes the bandicoot from other rats. Bandicoots have a robust form, a rounded head, large rounded or oval ears, and a short, broad muzzle. Their long and naked scaly tail is typical of practically all rats and mice. Bandicoots erect their piles of long hairs and grunt when excited. Bandicoots are found practically on the whole of the subcontinent from the Himalayas to Cape Comorin, including Sri Lanka, but they are not found in the deserts and the semi-arid zones of north-west India. Here, they are replaced by a related species, the short-tailed bandicoot (see section 3.1.2 below). The bandicoot is essentially parasitic on man, living in or about human dwellings. They cause a lot of damage to grounds and fl oorings because of their burrowing habits; they also dig tunnels through bricks and masonry. -
![Wild Mammals of the Annapurna Conservation Area Cggk"0F{ ;+/If0f If]Qsf :Tgwf/L Jgohgt' Wild Mammals of the Annapurna Conservation Area - 2019](https://docslib.b-cdn.net/cover/7316/wild-mammals-of-the-annapurna-conservation-area-cggk-0f-if0f-if-qsf-tgwf-l-jgohgt-wild-mammals-of-the-annapurna-conservation-area-2019-127316.webp)
Wild Mammals of the Annapurna Conservation Area Cggk"0F{ ;+/If0f If]Qsf :Tgwf/L Jgohgt' Wild Mammals of the Annapurna Conservation Area - 2019
Wild Mammals of the Annapurna Conservation Area cGgk"0f{ ;+/If0f If]qsf :tgwf/L jGohGt' Wild Mammals of the Annapurna Conservation Area - 2019 ISBN 978-9937-8522-8-9978-9937-8522-8-9 9 789937 852289 National Trust for Nature Conservation Annapurna Conservation Area Project Khumaltar, Lalitpur, Nepal Hariyo Kharka, Pokhara, Kaski, Nepal National Trust for Nature Conservation P.O. Box: 3712, Kathmandu, Nepal P.O. Box: 183, Kaski, Nepal Tel: +977-1-5526571, 5526573, Fax: +977-1-5526570 Tel: +977-61-431102, 430802, Fax: +977-61-431203 Annapurna Conservation Area Project Email: [email protected] Email: [email protected] Website: www.ntnc.org.np Website: www.ntnc.org.np 2019 Wild Mammals of the Annapurna Conservation Area cGgk"0f{ ;+/If0f If]qsf :tgwf/L jGohGt' National Trust for Nature Conservation Annapurna Conservation Area Project 2019 Wild Mammals of the Annapurna Conservation Area cGgk"0f{ ;+/If0f If]qsf :tgwf/L jGohGt' Published by © NTNC-ACAP, 2019 All rights reserved Any reproduction in full or in part must mention the title and credit NTNC-ACAP. Reviewers Prof. Karan Bahadur Shah (Himalayan Nature), Dr. Naresh Subedi (NTNC, Khumaltar), Dr. Will Duckworth (IUCN) and Yadav Ghimirey (Friends of Nature, Nepal). Compilers Rishi Baral, Ashok Subedi and Shailendra Kumar Yadav Suggested Citation Baral R., Subedi A. & Yadav S.K. (Compilers), 2019. Wild Mammals of the Annapurna Conservation Area. National Trust for Nature Conservation, Annapurna Conservation Area Project, Pokhara, Nepal. First Edition : 700 Copies ISBN : 978-9937-8522-8-9 Front Cover : Yellow-bellied Weasel (Mustela kathiah), back cover: Orange- bellied Himalayan Squirrel (Dremomys lokriah). -

Checklist of the Mammals of Indonesia
CHECKLIST OF THE MAMMALS OF INDONESIA Scientific, English, Indonesia Name and Distribution Area Table in Indonesia Including CITES, IUCN and Indonesian Category for Conservation i ii CHECKLIST OF THE MAMMALS OF INDONESIA Scientific, English, Indonesia Name and Distribution Area Table in Indonesia Including CITES, IUCN and Indonesian Category for Conservation By Ibnu Maryanto Maharadatunkamsi Anang Setiawan Achmadi Sigit Wiantoro Eko Sulistyadi Masaaki Yoneda Agustinus Suyanto Jito Sugardjito RESEARCH CENTER FOR BIOLOGY INDONESIAN INSTITUTE OF SCIENCES (LIPI) iii © 2019 RESEARCH CENTER FOR BIOLOGY, INDONESIAN INSTITUTE OF SCIENCES (LIPI) Cataloging in Publication Data. CHECKLIST OF THE MAMMALS OF INDONESIA: Scientific, English, Indonesia Name and Distribution Area Table in Indonesia Including CITES, IUCN and Indonesian Category for Conservation/ Ibnu Maryanto, Maharadatunkamsi, Anang Setiawan Achmadi, Sigit Wiantoro, Eko Sulistyadi, Masaaki Yoneda, Agustinus Suyanto, & Jito Sugardjito. ix+ 66 pp; 21 x 29,7 cm ISBN: 978-979-579-108-9 1. Checklist of mammals 2. Indonesia Cover Desain : Eko Harsono Photo : I. Maryanto Third Edition : December 2019 Published by: RESEARCH CENTER FOR BIOLOGY, INDONESIAN INSTITUTE OF SCIENCES (LIPI). Jl Raya Jakarta-Bogor, Km 46, Cibinong, Bogor, Jawa Barat 16911 Telp: 021-87907604/87907636; Fax: 021-87907612 Email: [email protected] . iv PREFACE TO THIRD EDITION This book is a third edition of checklist of the Mammals of Indonesia. The new edition provides remarkable information in several ways compare to the first and second editions, the remarks column contain the abbreviation of the specific island distributions, synonym and specific location. Thus, in this edition we are also corrected the distribution of some species including some new additional species in accordance with the discovery of new species in Indonesia. -

Mammal Species Native to the USA and Canada for Which the MIL Has an Image (296) 31 July 2021
Mammal species native to the USA and Canada for which the MIL has an image (296) 31 July 2021 ARTIODACTYLA (includes CETACEA) (38) ANTILOCAPRIDAE - pronghorns Antilocapra americana - Pronghorn BALAENIDAE - bowheads and right whales 1. Balaena mysticetus – Bowhead Whale BALAENOPTERIDAE -rorqual whales 1. Balaenoptera acutorostrata – Common Minke Whale 2. Balaenoptera borealis - Sei Whale 3. Balaenoptera brydei - Bryde’s Whale 4. Balaenoptera musculus - Blue Whale 5. Balaenoptera physalus - Fin Whale 6. Eschrichtius robustus - Gray Whale 7. Megaptera novaeangliae - Humpback Whale BOVIDAE - cattle, sheep, goats, and antelopes 1. Bos bison - American Bison 2. Oreamnos americanus - Mountain Goat 3. Ovibos moschatus - Muskox 4. Ovis canadensis - Bighorn Sheep 5. Ovis dalli - Thinhorn Sheep CERVIDAE - deer 1. Alces alces - Moose 2. Cervus canadensis - Wapiti (Elk) 3. Odocoileus hemionus - Mule Deer 4. Odocoileus virginianus - White-tailed Deer 5. Rangifer tarandus -Caribou DELPHINIDAE - ocean dolphins 1. Delphinus delphis - Common Dolphin 2. Globicephala macrorhynchus - Short-finned Pilot Whale 3. Grampus griseus - Risso's Dolphin 4. Lagenorhynchus albirostris - White-beaked Dolphin 5. Lissodelphis borealis - Northern Right-whale Dolphin 6. Orcinus orca - Killer Whale 7. Peponocephala electra - Melon-headed Whale 8. Pseudorca crassidens - False Killer Whale 9. Sagmatias obliquidens - Pacific White-sided Dolphin 10. Stenella coeruleoalba - Striped Dolphin 11. Stenella frontalis – Atlantic Spotted Dolphin 12. Steno bredanensis - Rough-toothed Dolphin 13. Tursiops truncatus - Common Bottlenose Dolphin MONODONTIDAE - narwhals, belugas 1. Delphinapterus leucas - Beluga 2. Monodon monoceros - Narwhal PHOCOENIDAE - porpoises 1. Phocoena phocoena - Harbor Porpoise 2. Phocoenoides dalli - Dall’s Porpoise PHYSETERIDAE - sperm whales Physeter macrocephalus – Sperm Whale TAYASSUIDAE - peccaries Dicotyles tajacu - Collared Peccary CARNIVORA (48) CANIDAE - dogs 1. Canis latrans - Coyote 2. -

Repositiorio | FAUBA | Artículos De Docentes E Investigadores De FAUBA
Biodivers Conserv (2011) 20:3077–3100 DOI 10.1007/s10531-011-0118-9 REVIEW PAPER Effects of agriculture expansion and intensification on the vertebrate and invertebrate diversity in the Pampas of Argentina Diego Medan • Juan Pablo Torretta • Karina Hodara • Elba B. de la Fuente • Norberto H. Montaldo Received: 23 July 2010 / Accepted: 15 July 2011 / Published online: 24 July 2011 Ó Springer Science+Business Media B.V. 2011 Abstract In this paper we summarize for the first time the effects of agriculture expansion and intensification on animal diversity in the Pampas of Argentina and discuss research needs for biodiversity conservation in the area. The Pampas experienced little human intervention until the last decades of the 19th century. Agriculture expanded quickly during the 20th century, transforming grasslands into cropland and pasture lands and converting the landscape into a mosaic of natural fragments, agricultural fields, and linear habitats. In the 1980s, agriculture intensification and replacement of cattle grazing- cropping systems by continuous cropping promoted a renewed homogenisation of the most productive areas. Birds and carnivores were more strongly affected than rodents and insects, but responses varied within groups: (a) the geographic ranges and/or abundances of many native species were reduced, including those of carnivores, herbivores, and specialist species (grassland-adapted birds and rodents, and probably specialized pollinators), sometimes leading to regional extinction (birds and large carnivores), (b) other native species were unaffected (birds) or benefited (bird, rodent and possibly generalist pollinator and crop-associated insect species), (c) novel species were introduced, thus increasing species richness of most groups (26% of non-rodent mammals, 11.1% of rodents, 6.2% of birds, 0.8% of pollinators). -

Controlled Animals
Environment and Sustainable Resource Development Fish and Wildlife Policy Division Controlled Animals Wildlife Regulation, Schedule 5, Part 1-4: Controlled Animals Subject to the Wildlife Act, a person must not be in possession of a wildlife or controlled animal unless authorized by a permit to do so, the animal was lawfully acquired, was lawfully exported from a jurisdiction outside of Alberta and was lawfully imported into Alberta. NOTES: 1 Animals listed in this Schedule, as a general rule, are described in the left hand column by reference to common or descriptive names and in the right hand column by reference to scientific names. But, in the event of any conflict as to the kind of animals that are listed, a scientific name in the right hand column prevails over the corresponding common or descriptive name in the left hand column. 2 Also included in this Schedule is any animal that is the hybrid offspring resulting from the crossing, whether before or after the commencement of this Schedule, of 2 animals at least one of which is or was an animal of a kind that is a controlled animal by virtue of this Schedule. 3 This Schedule excludes all wildlife animals, and therefore if a wildlife animal would, but for this Note, be included in this Schedule, it is hereby excluded from being a controlled animal. Part 1 Mammals (Class Mammalia) 1. AMERICAN OPOSSUMS (Family Didelphidae) Virginia Opossum Didelphis virginiana 2. SHREWS (Family Soricidae) Long-tailed Shrews Genus Sorex Arboreal Brown-toothed Shrew Episoriculus macrurus North American Least Shrew Cryptotis parva Old World Water Shrews Genus Neomys Ussuri White-toothed Shrew Crocidura lasiura Greater White-toothed Shrew Crocidura russula Siberian Shrew Crocidura sibirica Piebald Shrew Diplomesodon pulchellum 3. -
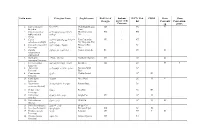
Red List of Endemic IUCN Red CITES Bern Bonn Georgia Species of the List Conventi Convention Caucasus on (CMS) 1
Latin name Georgian Name English name Red List of Endemic IUCN Red CITES Bern Bonn Georgia species of the list Conventi Convention Caucasus on (CMS) 1. Capra aegagrus niamori Wild Goat, Bezoar CR VU II Erxleben. Goat 2. Capra caucasica dasavleTkavkasiuri West Caucasian EN + EN Güldenstädt & jixvi Tur Pallas. 3. Capra aRmosavleTkavkasiuri East Caucasian VU + NT cylindricornis Blyth. jixvi Tur, Dagestan Tur 4. Capreolus capreolus evropuli Sveli European Roe LC Linnaeus. Deer 5. Gazella qurciki, jeirani Goitered Gazelle RE VU II subgutturosa Güldenstädt. 6. Rupicapra arCvi, fsiti Northern Chamois EN LC II rupicapra Linnaeus. 7. Cervus elaphus keTilSobili iremi Red Deer CR LC II I Linnaeus. 8. Sus scrofa gareuli Rori, taxi Eurasian Wild LC Linnaeus. Boar 9. Canis aureus tura Golden Jackal LC III Linnaeus. 10. Canis lupus mgeli Grey Wolf LC II II Linnaeus. 11. Nyctereutes enotisebri ZaRli Racoon Dog LC procyonoides Gray. 12. Vulpes vulpes mela Red Fox LC III Linnaeus. 13. Felis chaus lelianis kata Jungle Cat VU LC II Schreber. 14. Felis silvestris tyis kata Wild Cat LC II II Shreber. 15. Felis libyca Forster. velis kata Steppe Cat 16. Lynx lynx Linnaeus. focxveri Eurasian Lynx CR LC II 17. Panthera pardus jiqi Leopard CR NT I II Linnaeus. 18. Hyaena hyaena afTari Striped Hyaena CR NT Linnaeus. 19. Lutra lutra wavi Eurasian Otter, VU NT I II Linnaeus. Common Otter 20. Martes foina kldis kverna Stone Marten, LC III Erxleben. Beech Marten 21. Martes martes tyis kverna European Pine LC Linnaeus. Marten 22. Meles meles maCvi Eurasian Badger LC Linnaeus. 23. Mustela lutreola waula European Mink EN II Linnaeus. -
Checklist of Rodents and Insectivores of the Mordovia, Russia
ZooKeys 1004: 129–139 (2020) A peer-reviewed open-access journal doi: 10.3897/zookeys.1004.57359 RESEARCH ARTICLE https://zookeys.pensoft.net Launched to accelerate biodiversity research Checklist of rodents and insectivores of the Mordovia, Russia Alexey V. Andreychev1, Vyacheslav A. Kuznetsov1 1 Department of Zoology, National Research Mordovia State University, Bolshevistskaya Street, 68. 430005, Saransk, Russia Corresponding author: Alexey V. Andreychev ([email protected]) Academic editor: R. López-Antoñanzas | Received 7 August 2020 | Accepted 18 November 2020 | Published 16 December 2020 http://zoobank.org/C127F895-B27D-482E-AD2E-D8E4BDB9F332 Citation: Andreychev AV, Kuznetsov VA (2020) Checklist of rodents and insectivores of the Mordovia, Russia. ZooKeys 1004: 129–139. https://doi.org/10.3897/zookeys.1004.57359 Abstract A list of 40 species is presented of the rodents and insectivores collected during a 15-year period from the Republic of Mordovia. The dataset contains more than 24,000 records of rodent and insectivore species from 23 districts, including Saransk. A major part of the data set was obtained during expedition research and at the biological station. The work is based on the materials of our surveys of rodents and insectivo- rous mammals conducted in Mordovia using both trap lines and pitfall arrays using traditional methods. Keywords Insectivores, Mordovia, rodents, spatial distribution Introduction There is a need to review the species composition of rodents and insectivores in all regions of Russia, and the work by Tovpinets et al. (2020) on the Crimean Peninsula serves as an example of such research. Studies of rodent and insectivore diversity and distribution have a long history, but there are no lists for many regions of Russia of Copyright A.V. -

Molecular Phylogeny of Mobatviruses (Hantaviridae) in Myanmar and Vietnam
viruses Article Molecular Phylogeny of Mobatviruses (Hantaviridae) in Myanmar and Vietnam Satoru Arai 1, Fuka Kikuchi 1,2, Saw Bawm 3 , Nguyễn Trường Sơn 4,5, Kyaw San Lin 6, Vương Tân Tú 4,5, Keita Aoki 1,7, Kimiyuki Tsuchiya 8, Keiko Tanaka-Taya 1, Shigeru Morikawa 9, Kazunori Oishi 1 and Richard Yanagihara 10,* 1 Infectious Disease Surveillance Center, National Institute of Infectious Diseases, Tokyo 162-8640, Japan; [email protected] (S.A.); [email protected] (F.K.); [email protected] (K.A.); [email protected] (K.T.-T.); [email protected] (K.O.) 2 Department of Chemistry, Faculty of Science, Tokyo University of Science, Tokyo 162-8601, Japan 3 Department of Pharmacology and Parasitology, University of Veterinary Science, Yezin, Nay Pyi Taw 15013, Myanmar; [email protected] 4 Institute of Ecology and Biological Resources, Vietnam Academy of Science and Technology, Hanoi, Vietnam; [email protected] (N.T.S.); [email protected] (V.T.T.) 5 Graduate University of Science and Technology, Vietnam Academy of Science and Technology, Hanoi, Vietnam 6 Department of Aquaculture and Aquatic Disease, University of Veterinary Science, Yezin, Nay Pyi Taw 15013, Myanmar; [email protected] 7 Department of Liberal Arts, Faculty of Science, Tokyo University of Science, Tokyo 162-8601, Japan 8 Laboratory of Bioresources, Applied Biology Co., Ltd., Tokyo 107-0062, Japan; [email protected] 9 Department of Veterinary Science, National Institute of Infectious Diseases, Tokyo 162-8640, Japan; [email protected] 10 Pacific Center for Emerging Infectious Diseases Research, John A.