Human Cytochrome B5 Reductase: Structure, Function, and Potential Applications
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Characterization of a Microsomal Retinol Dehydrogenase Gene from Amphioxus: Retinoid Metabolism Before Vertebrates
Chemico-Biological Interactions 130–132 (2001) 359–370 www.elsevier.com/locate/chembiont Characterization of a microsomal retinol dehydrogenase gene from amphioxus: retinoid metabolism before vertebrates Diana Dalfo´, Cristian Can˜estro, Ricard Albalat, Roser Gonza`lez-Duarte * Departament de Gene`tica, Facultat de Biologia, Uni6ersitat de Barcelona, A6. Diagonal, 645, E-08028, Barcelona, Spain Abstract Amphioxus, a member of the subphylum Cephalochordata, is thought to be the closest living relative to vertebrates. Although these animals have a vertebrate-like response to retinoic acid, the pathway of retinoid metabolism remains unknown. Two different enzyme systems — the short chain dehydrogenase/reductases and the cytosolic medium-chain alcohol dehydrogenases (ADHs) — have been postulated in vertebrates. Nevertheless, recent data show that the vertebrate-ADH1 and ADH4 retinol-active forms originated after the divergence of cephalochordates and vertebrates. Moreover, no data has been gathered in support of medium-chain retinol active forms in amphioxus. Then, if the cytosolic ADH system is absent and these animals use retinol, the microsomal retinol dehydrogenases could be involved in retinol oxidation. We have identified the genomic region and cDNA of an amphioxus Rdh gene as a preliminary step for functional characterization. Besides, phyloge- netic analysis supports the ancestral position of amphioxus Rdh in relation to the vertebrate forms. © 2001 Elsevier Science Ireland Ltd. All rights reserved. Keywords: Retinol dehydrogenase; Retinoid metabolism; Amphioxus * Corresponding author. Fax: +34-93-4110969. E-mail address: [email protected] (R. Gonza`lez-Duarte). 0009-2797/01/$ - see front matter © 2001 Elsevier Science Ireland Ltd. All rights reserved. PII: S0009-2797(00)00261-1 360 D. -

SUPPLEMENTARY DATA Data Supplement To
SUPPLEMENTARY DATA Data supplement to Perivascular adipose tissue controls insulin-stimulated perfusion, mitochondrial protein expression and glucose uptake in muscle through adipomuscular microvascular anastomoses Surname first author Turaihi Authors & Affiliations Turaihi, Alexander H MD1; Serné, Erik H, MD PhD2; Molthoff, Carla FM PhD3; Koning, Jasper J PhD4; Knol, Jaco PhD6; Niessen, Hans W MD PhD5; Goumans, Marie Jose TH PhD7; van Poelgeest, Erik M MD1; Yudkin, John S MD PhD8; Smulders, Yvo M MD PhD2; Connie R Jimenez, PhD6; van Hinsbergh, Victor WM PhD1; Eringa, Etto C PhD1 ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db18-1066/-/DC1 SUPPLEMENTARY DATA Western immunoblotting Skeletal muscle samples were lysed up in 1D‐sample buffer (10% glycerol, 62.5 mmol/L Tris (pH 6.8), 2% w/v LDS, 2% w/v DTT) and protein concentration was determined using Pierce 660‐nm protein assay (Thermo scientific, Waltham, MA USA 02 451; 22 660) according to the manufacturer's instructions. Heat shock protein 90 immunoblotting was performed by application of samples (5 µg protein) on 4‐15% Criterion TGX gels (Biorad, Veenendaal, the Netherlands, 5 671 084) and semi‐dry blotting onto PVDF membranes (GE Healthcare‐Fisher, RPN1416F), incubated overnight with rat monoclonal HSP90 antibody (1:1000 dilution) after blocking with 5% milk in TBS‐T (137 mM NaCl, 20 mmol/L Tris pH 7.0 and 0.1% (v/v) Tween [Sigma‐Aldrich, P7949]). After 2 hours incubation with anti-rat, horse radish peroxidase-coupled secondary antibody (Thermo Fisher 62-9520), the blot was stained using ECL‐prime (Fisher scientific, 10 308 449) and analysed on an AI‐600 imaging system (GE Healthcare, Life Sciences). -

Table 1. Identified Proteins with Expression Significantly Altered in the Hippocampus of Rats of Exposed Group (Pb) Vs
Table 1. Identified proteins with expression significantly altered in the hippocampus of rats of exposed group (Pb) vs. Control. Fold Change Accession Id a Protein Description Score Pb P35213 14-3-3 protein beta/alpha 85420 −0.835 P62260 14-3-3 protein epsilon 96570 −0.878 P68511 14-3-3 protein eta 85420 −0.844 P68255 14-3-3 protein theta 85420 −0.835 P63102 14-3-3 protein zeta/delta 105051 −0.803 P13233 2',3'-cyclic-nucleotide 3'-phosphodiesterase 151400 1.405 P68035 Actin, alpha cardiac muscle 1 442584 −0.942 P68136 Actin, alpha skeletal muscle 441060 −0.970 P62738 Actin, aortic smooth muscle 438270 −0.970 P60711 Actin, cytoplasmic 1 630104 −0.942 P63259 Actin, cytoplasmic 2 630104 −0.942 P63269 Actin, gamma-enteric smooth muscle 438270 −0.951 Q05962 ADP/ATP translocase 1 60100 −0.554 Q09073 ADP/ATP translocase 2 49102 −0.482 P84079 ADP-ribosylation factor 1 34675 −0.644 P84082 ADP-ribosylation factor 2 22412 −0.644 P61206 ADP-ribosylation factor 3 34675 −0.619 P61751 ADP-ribosylation factor 4 22412 −0.670 P84083 ADP-ribosylation factor 5 22412 −0.625 P04764 Alpha-enolase 46219 −0.951 P23565 Alpha-internexin 9478 1.062 P37377 Alpha-synuclein 89619 −0.771 P13221 Aspartate aminotransferase, cytoplasmic 23661 1.083 P00507 Aspartate aminotransferase, mitochondrial 46049 1.116 P10719 ATP synthase subunit beta, mitochondrial 232442 −0.835 P85969 Beta-soluble NSF attachment protein 9638 1.419 Q63754 Beta-synuclein 66842 −0.779 P11275 Calcium/calmodulin-dependent protein kinase type II subunit alpha 181954 1.105 P08413 Calcium/calmodulin-dependent protein kinase type II subunit beta 80840 1.127 P15791 Calcium/calmodulin-dependent protein kinase type II subunit delta 62682 1.105 Int. -

Methionine Sulfoxide Reductase a Is a Stereospecific Methionine Oxidase
Methionine sulfoxide reductase A is a stereospecific methionine oxidase Jung Chae Lim, Zheng You, Geumsoo Kim, and Rodney L. Levine1 Laboratory of Biochemistry, National Heart, Lung, and Blood Institute, Bethesda, MD 20892-8012 Edited by Irwin Fridovich, Duke University Medical Center, Durham, NC, and approved May 10, 2011 (received for review February 10, 2011) Methionine sulfoxide reductase A (MsrA) catalyzes the reduction Results of methionine sulfoxide to methionine and is specific for the S epi- Stoichiometry. Branlant and coworkers have studied in careful mer of methionine sulfoxide. The enzyme participates in defense detail the mechanism of the MsrA reaction in bacteria (17, 18). against oxidative stresses by reducing methionine sulfoxide resi- In the absence of reducing agents, each molecule of MsrA dues in proteins back to methionine. Because oxidation of methio- reduces two molecules of MetO. Reduction of the first MetO nine residues is reversible, this covalent modification could also generates a sulfenic acid at the active site cysteine, and it is function as a mechanism for cellular regulation, provided there reduced back to the thiol by a fast reaction, which generates a exists a stereospecific methionine oxidase. We show that MsrA disulfide bond in the resolving domain of the protein. The second itself is a stereospecific methionine oxidase, producing S-methio- MetO is then reduced and again generates a sulfenic acid at the nine sulfoxide as its product. MsrA catalyzes its own autooxidation active site. Because the resolving domain cysteines have already as well as oxidation of free methionine and methionine residues formed a disulfide, no further reaction forms. -

The Flavodoxin Flda Activates the Class Ia Ribonucleotide Reductase of Campylobacter Jejuni
This is a repository copy of The flavodoxin FldA activates the class Ia ribonucleotide reductase of Campylobacter jejuni. White Rose Research Online URL for this paper: http://eprints.whiterose.ac.uk/173101/ Version: Published Version Article: Alqurashi, A., Alfs, L., Swann, J. et al. (2 more authors) (2021) The flavodoxin FldA activates the class Ia ribonucleotide reductase of Campylobacter jejuni. Molecular Microbiology. ISSN 0950-382X https://doi.org/10.1111/mmi.14715 Reuse This article is distributed under the terms of the Creative Commons Attribution (CC BY) licence. This licence allows you to distribute, remix, tweak, and build upon the work, even commercially, as long as you credit the authors for the original work. More information and the full terms of the licence here: https://creativecommons.org/licenses/ Takedown If you consider content in White Rose Research Online to be in breach of UK law, please notify us by emailing [email protected] including the URL of the record and the reason for the withdrawal request. [email protected] https://eprints.whiterose.ac.uk/ Received: 29 October 2020 | Revised: 11 March 2021 DOI: 10.1111/mmi.14715 RESEARCH ARTICLE The flavodoxin FldA activates the class Ia ribonucleotide reductase of Campylobacter jejuni Abdulmajeed Alqurashi1 | Laura Alfs2 | Jordan Swann2 | Julea N. Butt2 | David J. Kelly 1 1Department of Molecular Biology and Biotechnology, The University of Sheffield, Abstract Sheffield, UK Campylobacter jejuni is a microaerophilic zoonotic pathogen with an atypical res- 2 School of Chemistry, University of East piratory Complex I that oxidizes a flavodoxin (FldA) instead of NADH. FldA is es- Anglia, Norwich, UK sential for viability and is reduced via pyruvate and 2- oxoglutarate oxidoreductases Correspondence (POR/OOR). -

Iron Deficiency in the Rat: Effects on Neutrophil Activation and Metabolism
THEOPHYLLINE AND BRAIN 549 A dietary protein deficiency can affect the susceptibility to the metabolism in children with protein-calorie malnutrition. Am. J. Clin. Nutr., 28: 977 (1975). toxicity of drugs or other agents such as pesticides and herbicides 8. Nakamoto. T. and Miller. S. A.: Effect of vrotein-energy malnutrition on the (2). At the present time, there are no such studies relative to growth df mandible aid long bone in newborn miie and female rats. J. theophylline, although the usage of theophylline is now quite Nutr., 107: 983 (1977). common in the neonatal intensive care environment. Many 9. Nebron, R. M., Resnick, M. D., and Halstmm, W. J.: Developmental outcome of premature infants treated with theophylline. Dev. Pharmacol. Ther., 1: infants therein who are now receiving theophylline therapeuti- 274 ( 1980). cally are being dosed on a body weight basis without considera- 10. Newberne, P. M., Gross, R. L., and Roe, D. A,: Dmg, toxin, nutrient interac- tion of the nutritional status. Our data suggest that, in the animal tions. World Rev. Nutr. Diet., 29: 130 (1978). model, the administration of theophylline in the presence of a 1 I. Pastorova, B., Sova, O., and Burda, J.: Incorporation of I4C-thymidine into liver and brain DNA of protein-deficient rats. Physiol. Bohemoslov., 27: 69 compromised nutritional status may have effects not now appar- (1978). ent. We add our concern to that expressed by others (16) that 12. Prasad, A. S., Dumouchell, E., Kovich, D., and Oberleas, D.: A simple methylxanthine administration may have previously unsus- fluorometric method for the determination of RNA and DNA in tissue. -

CYB5R3 Gene Cytochrome B5 Reductase 3
CYB5R3 gene cytochrome b5 reductase 3 Normal Function The CYB5R3 gene provides instruction for making an enzyme called cytochrome b5 reductase 3. This enzyme is involved in transferring negatively charged particles called electrons from one molecule to another. Two versions (isoforms) of this enzyme are produced from the CYB5R3 gene. The soluble isoform is present only in red blood cells, and the membrane-bound isoform is found in all other cell types. Normal red blood cells contain molecules of iron-containing hemoglobin, which deliver oxygen to the body's tissues. The iron in hemoglobin is ferrous (Fe2+), but it can spontaneously become ferric (Fe3+). Hemoglobin that contains ferric iron is called methemoglobin, and it cannot deliver oxygen. The soluble isoform of cytochrome b5 reductase 3 changes ferric iron back to ferrous iron so hemoglobin can function. Normally, red blood cells contain less than 2 percent methemoglobin. The membrane-bound isoform is embedded in the membranes of various cellular compartments and is widely used in the body. This isoform is necessary for many chemical reactions, including the breakdown and formation of fatty acids, the formation of cholesterol, and the breakdown of various molecules and drugs. Health Conditions Related to Genetic Changes Autosomal recessive congenital methemoglobinemia More than 65 mutations in the CYB5R3 gene have been found to cause autosomal recessive congenital methemoglobinemia types I and II. Most of these CYB5R3 gene mutations cause autosomal recessive congenital methemoglobinemia type I, which is characterized by a lack of oxygen in the body's tissues and bluish appearance of the skin, lips, and nails (cyanosis). -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Mutations in SDHD Lead to Autosomal Recessive Encephalomyopathy And
Downloaded from http://jmg.bmj.com/ on December 5, 2017 - Published by group.bmj.com Genotype-phenotype correlations ORIGINAL ARTICLE Mutations in SDHD lead to autosomal recessive encephalomyopathy and isolated mitochondrial complex II deficiency Christopher Benjamin Jackson,1,2 Jean-Marc Nuoffer,3 Dagmar Hahn,3 Holger Prokisch,4,5 Birgit Haberberger,4 Matthias Gautschi,3,6 Annemarie Häberli,3 Sabina Gallati,1 André Schaller1 ▸ Additional material is ABSTRACT succinate to fumarate, is formed by the two larger published online only. To view Background Defects of the mitochondrial respiratory hydrophilic subunits, SDHA and SDHB, which also please visit the journal online (10.1136/jmedgenet-2013- chain complex II (succinate dehydrogenase (SDH) harbour the redox cofactors that participate in elec- 101932) complex) are extremely rare. Of the four nuclear encoded tron transfer to ubiquinone. The cofactor FAD is 1 proteins composing complex II, only mutations in the covalently bound to SDHA which provides the suc- Division of Human Genetics, fl fi Departments of Paediatrics and 70 kDa avoprotein (SDHA) and the recently identi ed cinate binding site, and SDHB possesses three Fe-S Clinical Research, University of complex II assembly factor (SDHAF1) have been found to centres which mediate the electron transfer to ubi- Bern, Bern, Switzerland be causative for mitochondrial respiratory chain diseases. quinone. The smaller hydrophobic SDHC and 2 Graduate School for Cellular Mutations in the other three subunits (SDHB, SDHC, SDHD subunits constitute the membrane anchor and Biomedical Sciences, SDHD) and the second assembly factor (SDHAF2) have and ubiquinone binding sites of CII and localise University of Bern, Bern, 2–4 Switzerland so far only been associated with hereditary CII to the inner mitochondrial membrane. -

Chain of Human Neutrophil Cytochrome B CHARLES A
Proc. Nati. Acad. Sci. USA Vol. 85, pp. 3319-3323, May 1988 Biochemistry Primary structure and unique expression of the 22-kilodalton light chain of human neutrophil cytochrome b CHARLES A. PARKOS*, MARY C. DINAUERt, LESLIE E. WALKER*, RODGER A. ALLEN*, ALGIRDAS J. JESAITIS*, AND STUART H. ORKINtt *Department of Immunology, Research Institute of the Scripps Clinic, La Jolla, CA 92037; tDivision of Hematology-Oncology, Children's Hospital, and Dana-Farber Cancer Institute, Department of Pediatrics, Harvard Medical School, Boston, MA 02115; and tHoward Hughes Medical Institute, Children's Hospital, Boston, MA 02115 Communicated by Harvey F. Lodish, January 14, 1988 ABSTRACT Cytochrome b comprising 91-kDa and 22- Cytochrome b purified from neutrophil membranes ap- kDa subunits is a critical component of the membrane-bound pears to be a heterodimer of a glycosylated 91-kDa heavy oxidase of phagocytes that generates superoxide. This impor- chain and a nonglycosylated 22-kDa light chain (10-12). The tant microbicidal system is impaired in inherited disorders 91-kDa subunit is encoded by a gene designated CGD, known as chronic granulomatous disease (CGD). Previously we residing at chromosomal position Xp2l, which originally was determined the sequence of the larger subunit from the cDNA identified on the basis of genetic linkage without reference to of the CGD gene, the X chromosome locus affected in "X- a specific protein product (8). Antisera generated to either a linked" CGD. To complete the primary structure of the synthetic peptide predicted from the cDNA or to a fusion cytochrome b and to assess expression of the smaller subunit, protein produced in E. -

Electron Transport Discovery Four Complexes Complex I: Nadhà Coqh2
BI/CH 422/622 OUTLINE: Pyruvate pyruvate dehydrogenase Krebs’ Cycle How did he figure it out? Overview 8 Steps Citrate Synthase Aconitase Isocitrate dehydrogenase Ketoglutarate dehydrogenase Succinyl-CoA synthetase Succinate dehydrogenase Fumarase Malate dehydrogenase Energetics Regulation Summary Oxidative Phosphorylation Energetics (–0.16 V needed for making ATP) Mitochondria Transport (2.4 kcal/mol needed to transport H+ out) Electron transport Discovery Four Complexes Complex I: NADHà CoQH2 Complex II: Succinateà CoQH2 2+ Complex III: CoQH2à Cytochrome C (Fe ) 2+ Complex IV: Cytochrome C (Fe ) à H2O Electron Transport à O2 Inhibitors of Electron Transport Big Drop! • Inhibitors all stop ET and ATP synthesis: very toxic! Spectral work Big Drop! NADH Cyto-a3 Cyto-c1 Big Drop! Cyto-b Cyto-c Cyto-a Fully reduced Flavin Cyto-c + rotenone + antimycin A 300 350 400 450 500 550 600 650 700 1 Electron Transport Electron-Transport Chain Complexes Contain a Series of Electron Carriers • Better techniques for isolating and handling mitochondria, and isolated various fractions of the inner mitochondrial membrane • Measure E°’ • They corresponded to these large drops, and they contained the redox compounds isolated previously. • When assayed for what reactions they could perform, they could perform certain redox reactions and not others. • When isolated, including isolating the individual redox compounds, and measuring the E°’ for each, it was clear that an electron chain was occurring; like a wire! • Lastly, when certain inhibitors were added, some of the redox reactions could be inhibited and others not. Site of the inhibition could be mapped. Electron Transport Electron-Transport Chain Complexes Contain a Series of Electron Carriers • Better techniques for isolating and handling mitochondria, and isolated various fractions of the inner mitochondrial membrane • Measure E°’ • They corresponded to these large drops, and they contained the redox compounds isolated previously. -
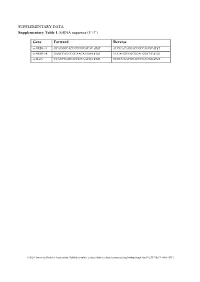
Supplementary Tables and Figures
SUPPLEMENTARY DATA Supplementary Table 1. SiRNA sequence (5’-3’) Gene Forward Reverse si-HRD1-1# GCAUGGCAGUCCUGUACAU dTdT AUGUACAGGACUGCCAUGC dTdT si-HRD1-2# GAGCCAUCCGCAACAUGAA dTdT UUCAUGUUGCGGAUGGCUC dTdT si-MafA CCAUCGAGUACGUCAACGA dTdT UCGUUGACGUACUCGAUGG dTdT ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 2. Primer sequences for qRT-PCR (5’-3’) Gene Forward Reverse human HRD1 GCTCACGCCTACTACCTCAAA GCCAGACAAGTCTCTGTGACG mouse mafA AAGCGGCGCACGCTCAAGAA GGTCCCGCTCCTTGGCCAGA mouse insulin1 CACTTCCTACCCCTGCTGG ACCACAAAGATGCTGTTTGACA mouse β-actin AGGCCAACCGTGAAAAGATG AGAGCATAGCCCTCGTAGATGG human β-actin CATGTACGTTGCTATCCAGGC CTCCTTAATGTCACGCACGAT ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 3. Primer sequences for ChIP (5’-3’) Gene promoter Forward Reverse mouse Insulin1, 2 GGAACTGTGAAACAGTCCAAGG CCCCCTGGACTTTGCTGTTTG ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 4. Primer sequences for PCR (5’-3’) Gene Forward Reverse HRD1-pDsred CCCAAGCTTATGTTCCGCACCGCAGT GGGGTACCCAGTGGGCAACAGGGG HRD1-pCMV- Flag GGGGTACCATGTTCCGCACCGCAGT CCCAAGCTTGTGGGCAACAGGGGACT C HRD1-pCMV-HA GGCCATGGGCCATATGGGATCCTTCC AGGGATGCCACCCGGGGATCCTCAGT GCACCGCAGTGATG GGGCAACAGGGGAC HRD1-N-HA GGCCATGGGCCATATGGGATCCTTCC