An Introduction to Scientific Image Data Analysis Using R
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Bioimage Analysis Tools
Bioimage Analysis Tools Kota Miura, Sébastien Tosi, Christoph Möhl, Chong Zhang, Perrine Paul-Gilloteaux, Ulrike Schulze, Simon Norrelykke, Christian Tischer, Thomas Pengo To cite this version: Kota Miura, Sébastien Tosi, Christoph Möhl, Chong Zhang, Perrine Paul-Gilloteaux, et al.. Bioimage Analysis Tools. Kota Miura. Bioimage Data Analysis, Wiley-VCH, 2016, 978-3-527-80092-6. hal- 02910986 HAL Id: hal-02910986 https://hal.archives-ouvertes.fr/hal-02910986 Submitted on 3 Aug 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. 2 Bioimage Analysis Tools 1 2 3 4 5 6 Kota Miura, Sébastien Tosi, Christoph Möhl, Chong Zhang, Perrine Pau/-Gilloteaux, - Ulrike Schulze,7 Simon F. Nerrelykke,8 Christian Tischer,9 and Thomas Penqo'" 1 European Molecular Biology Laboratory, Meyerhofstraße 1, 69117 Heidelberg, Germany National Institute of Basic Biology, Okazaki, 444-8585, Japan 2/nstitute for Research in Biomedicine ORB Barcelona), Advanced Digital Microscopy, Parc Científic de Barcelona, dBaldiri Reixac 1 O, 08028 Barcelona, Spain 3German Center of Neurodegenerative -

Sweave User Manual
Sweave User Manual Friedrich Leisch and R-core June 30, 2017 1 Introduction Sweave provides a flexible framework for mixing text and R code for automatic document generation. A single source file contains both documentation text and R code, which are then woven into a final document containing • the documentation text together with • the R code and/or • the output of the code (text, graphs) This allows the re-generation of a report if the input data change and documents the code to reproduce the analysis in the same file that contains the report. The R code of the complete 1 analysis is embedded into a LATEX document using the noweb syntax (Ramsey, 1998) which is usually used for literate programming Knuth(1984). Hence, the full power of LATEX (for high- quality typesetting) and R (for data analysis) can be used simultaneously. See Leisch(2002) and references therein for more general thoughts on dynamic report generation and pointers to other systems. Sweave uses a modular concept using different drivers for the actual translations. Obviously different drivers are needed for different text markup languages (LATEX, HTML, . ). Several packages on CRAN provide support for other word processing systems (see Appendix A). 2 Noweb files noweb (Ramsey, 1998) is a simple literate-programming tool which allows combining program source code and the corresponding documentation into a single file. A noweb file is a simple text file which consists of a sequence of code and documentation segments, called chunks: Documentation chunks start with a line that has an at sign (@) as first character, followed by a space or newline character. -

Rkward: a Comprehensive Graphical User Interface and Integrated Development Environment for Statistical Analysis with R
JSS Journal of Statistical Software June 2012, Volume 49, Issue 9. http://www.jstatsoft.org/ RKWard: A Comprehensive Graphical User Interface and Integrated Development Environment for Statistical Analysis with R Stefan R¨odiger Thomas Friedrichsmeier Charit´e-Universit¨atsmedizin Berlin Ruhr-University Bochum Prasenjit Kapat Meik Michalke The Ohio State University Heinrich Heine University Dusseldorf¨ Abstract R is a free open-source implementation of the S statistical computing language and programming environment. The current status of R is a command line driven interface with no advanced cross-platform graphical user interface (GUI), but it includes tools for building such. Over the past years, proprietary and non-proprietary GUI solutions have emerged, based on internal or external tool kits, with different scopes and technological concepts. For example, Rgui.exe and Rgui.app have become the de facto GUI on the Microsoft Windows and Mac OS X platforms, respectively, for most users. In this paper we discuss RKWard which aims to be both a comprehensive GUI and an integrated devel- opment environment for R. RKWard is based on the KDE software libraries. Statistical procedures and plots are implemented using an extendable plugin architecture based on ECMAScript (JavaScript), R, and XML. RKWard provides an excellent tool to manage different types of data objects; even allowing for seamless editing of certain types. The objective of RKWard is to provide a portable and extensible R interface for both basic and advanced statistical and graphical analysis, while not compromising on flexibility and modularity of the R programming environment itself. Keywords: GUI, integrated development environment, plugin, R. -

ESS — Emacs Speaks Statistics
ESS — Emacs Speaks Statistics ESS version 5.14 The ESS Developers (A.J. Rossini, R.M. Heiberger, K. Hornik, M. Maechler, R.A. Sparapani, S.J. Eglen, S.P. Luque and H. Redestig) Current Documentation by The ESS Developers Copyright c 2002–2010 The ESS Developers Copyright c 1996–2001 A.J. Rossini Original Documentation by David M. Smith Copyright c 1992–1995 David M. Smith Permission is granted to make and distribute verbatim copies of this manual provided the copyright notice and this permission notice are preserved on all copies. Permission is granted to copy and distribute modified versions of this manual under the conditions for verbatim copying, provided that the entire resulting derived work is distributed under the terms of a permission notice identical to this one. Chapter 1: Introduction to ESS 1 1 Introduction to ESS The S family (S, Splus and R) and SAS statistical analysis packages provide sophisticated statistical and graphical routines for manipulating data. Emacs Speaks Statistics (ESS) is based on the merger of two pre-cursors, S-mode and SAS-mode, which provided support for the S family and SAS respectively. Later on, Stata-mode was also incorporated. ESS provides a common, generic, and useful interface, through emacs, to many statistical packages. It currently supports the S family, SAS, BUGS/JAGS, Stata and XLisp-Stat with the level of support roughly in that order. A bit of notation before we begin. emacs refers to both GNU Emacs by the Free Software Foundation, as well as XEmacs by the XEmacs Project. The emacs major mode ESS[language], where language can take values such as S, SAS, or XLS. -

Species' Traits and Phylogenetic
Northern Michigan University NMU Commons All NMU Master's Theses Student Works 8-2018 CLIMATE DRIVEN RANGE SHIFTS OF NORTH AMERICAN SMALL MAMMALS: SPECIES’ TRAITS AND PHYLOGENETIC INFLUENCES Katie Nehiba [email protected] Follow this and additional works at: https://commons.nmu.edu/theses Part of the Ecology and Evolutionary Biology Commons Recommended Citation Nehiba, Katie, "CLIMATE DRIVEN RANGE SHIFTS OF NORTH AMERICAN SMALL MAMMALS: SPECIES’ TRAITS AND PHYLOGENETIC INFLUENCES" (2018). All NMU Master's Theses. 557. https://commons.nmu.edu/theses/557 This Open Access is brought to you for free and open access by the Student Works at NMU Commons. It has been accepted for inclusion in All NMU Master's Theses by an authorized administrator of NMU Commons. For more information, please contact [email protected],[email protected]. CLIMATE DRIVEN RANGE SHIFTS OF NORTH AMERICAN SMALL MAMMALS: SPECIES’ TRAITS AND PHYLOGENETIC INFLUENCES By Katie R. Nehiba THESIS Submitted to Northern Michigan University In partial fulfillment of the requirements For the degree of MASTER OF SCIENCE Office of Graduate Education and Research July 2018 ABSTRACT By Katie R. Nehiba Current anthropogenically-driven climate change is accelerating at an unprecedented rate. In response, species’ ranges may shift, tracking optimal climatic conditions. Species-specific differences may produce predictable differences in the extent of range shifts. I evaluated if patterns of predicted responses to climate change were strongly related to species’ taxonomic identities and/or ecological characteristics of species’ niches, elevation and precipitation. I evaluated differences in predicted range shifts in well-sampled small mammals that are restricted to North America: kangaroo rats, voles, chipmunks, and ground squirrels. -

Deducer: a Data Analysis GUI for R
JSS Journal of Statistical Software June 2012, Volume 49, Issue 8. http://www.jstatsoft.org/ Deducer: A Data Analysis GUI for R Ian Fellows University of California, Los Angeles Abstract While R has proven itself to be a powerful and flexible tool for data exploration and analysis, it lacks the ease of use present in other software such as SPSS and Minitab. An easy to use graphical user interface (GUI) can help new users accomplish tasks that would otherwise be out of their reach, and improves the efficiency of expert users by replacing fifty key strokes with five mouse clicks. With this in mind, Deducer presents dialogs that are understandable for the beginner, and yet contain all (or most) of the options that an experienced statistician, performing the same task, would want. An Excel-like spreadsheet is included for easy data viewing and editing. Deducer is based on Java's Swing GUI library and can be used on any common operating system. The GUI is independent of the specific R console and can easily be used by calling a text-based menu system. Graphical menus are provided for the JGR console and the Windows R GUI. Keywords: GUI, R. 1. Introduction R (R Development Core Team 2012) is a powerful statistical programming language that places the latest statistical techniques at one's fingertips through thousands of add-on packages available on the Comprehensive R Archive Network (CRAN) download servers. The price for all of this power is complexity. Because R analyses must be called as text commands, the user is required to find out the name of the function that will accomplish their task, and then remember that name along with the names of the variables to feed it, and its argument options. -

Reproducible Research.. Using Sweave, Knitr and Pandoc
Reproducible Research.. Using Sweave, Knitr and Pandoc Aedín Culhane [email protected] Nov 20th 2012 My R Course Website http://bcb.dfci.harvard.edu/~aedin/ My HSPH homepage http://www.hsph.harvard.edu/research/aedin-culhane/ When issues of reproducibility arise • ``Remember that microarray analysis you did six months ago? We ran a few more arrays. Can you add them to the project and repeat the same analysis?'' • ``The statistical analyst who looked at the data I generated previously is no longer available. Can you get someone else to analyze my new data set using the same methods (and thus producing a report I can expect to understand)?'' • ``Please write/edit the methods sections for the abstract/paper/grant proposal I am submitting based on the analysis you did several months ago.'' From Keith Baggerly • Selected articles published in Nature Genetics between January 2005 and December 2006 that had used profiling with microarrays • Of the 56 items retrieved electronically, 20 articles were considered potentially eligible for the project • The four teams were from – University of Alabama at Birmingham (UAB) – Stanford/Dana-Farber (SD) – London (L) and Ioannina/Trento (IT) • Each team was comprised of 3-6 scientists who worked together to evaluate each article. Results • Result could be reproduced n=2 • Reproduced with discrepancy n=6 • Could not be reproduced n=10 – No data n=4 (no data n=2, subset n=1, no reporter data n=1) – Confusion over matching of data to analysis (n=2) – Specialized software required and not available (n=1)m – Raw data available but could not be processed n=2 Reproducibility of Analysis Ioannidis JP, Allison DB, Ball CA, Coulibaly I, Cui X, Culhane AC, et al, (2009) Repeatability of published microarray gene expression Analyses. -

Thesis Entire.Pdf (12.90Mb)
Faculty OF Science AND TECHNOLOGY Department OF Computer Science TOWARD ReprODUCIBLE Analysis AND ExplorATION OF High-ThrOUGHPUT Biological Datasets — Bjørn Fjukstad A DISSERTATION FOR THE DEGREE OF Philosophiae Doctor – 2018 This thesis document was typeset using the UiT Thesis LaTEX Template. © 2018 – http://github.com/egraff/uit-thesis “Ta aldri problemene på forskudd, for da får du dem to ganger, men ta gjerne seieren på forskudd, for hvis ikke er det altfor sjelden du får oppleve den.” –Ivar Tollefsen AbstrACT There is a rapid growth in the number of available biological datasets due to the advent of high-throughput data collection instruments combined with cheap compute infrastructure. Modern instruments enable the analysis of biological data at different levels, from small DNA sequences through larger cell structures, and up to the function of entire organs. These new datasets have brought the need to develop new software packages to enable novel insights into the underlying biological mechanisms in the development and progression of diseases such as cancer. The heterogeneity of biological datasets require researchers to tailor the explo- ration and analyses with a wide range of different tools and systems. However, despite the need for their integration, few of them provide standard inter- faces for analyses implemented using different programming languages and frameworks. In addition, because of the many tools, different input parame- ters, and references to databases, it is necessary to record these correctly. The lack of such details complicates reproducing the original results and the reuse of the analyses on new datasets. This increases the analysis time and leaves unrealized potential for scientific insights. -
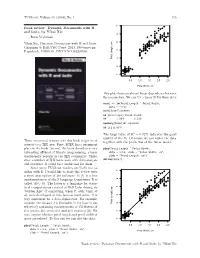
Dynamic Documents with R and Knitr, by Yihui Xie 116 Tugboat, Volume 35 (2014), No
TUGboat, Volume 35 (2014), No. 1 115 ● ● ● Book review: Dynamic Documents with R ● ● ● and knitr, by Yihui Xie ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● Boris Veytsman ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● Yihui Xie, Dynamic Documents with R and knitr. ● ● ● ● ● ● ● ● ● ● ● ● ● ● Chapman & Hall/CRC Press, 2013, 190+xxvi pp. ● ● ● ● ● ● ● ● ● US$ ISBN ● Paperback, 59.95. 978-1482203530. ● ● ● ● Petal Length, cm Petal ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● 1 2 3 4 5 6 7 0.5 1.0 1.5 2.0 2.5 Petal Width, cm This plot shows an almost linear dependence between the parameters. We can try a linear fit for these data: model <- lm(Petal.Length ˜ Petal.Width, data = iris) model$coefficients ## (Intercept) Petal.Width ## 1.084 2.230 summary(model)$r.squared ## [1] 0.9271 The large value of R2 = 0.9271 indicates the good quality of the fit. Of course we can replot the data There are several reasons why this book might be of together with the prediction of the linear model: interest to a TEX user. First, LATEX has a prominent place in the book. Second, the book describes a very plot(Petal.Length ˜ Petal.Width, interesting offshoot of literate programming, a topic data = iris, xlab = "Petal Width, cm", traditionally popular in the TEX community. Third, ylab = "Petal Length, cm") abline(model) since a number of TEX users work with data analysis and statistics, R could be a useful tool for them. Since some TUGboat readers are likely not fa- ● ● ● miliar with R, I would like to start this review with ● ● a short description of the software. R [1] is a free ● ● ● ● ● ● ● ● ● ● ● implementation of the S language (sometimes R is ● ● ● ● ● ● ● ● ● ● ● ● ● ● called GNU S). -

An Introduction to R Franz X
An Introduction to R Franz X. Mohr December, 2019 Contents 1 Introduction 2 2 Installation 2 2.1 R .................................... 3 2.2 A development environment: RStudio ................ 4 3 R(eal) Basics 5 3.1 Functions and packages ........................ 5 3.2 Before you start ............................ 7 3.3 Getting help .............................. 8 3.4 Importing data ............................. 9 4 Data transformation 12 4.1 Standard method ........................... 12 4.2 Manipulating data with the dplyr package ............. 15 5 Saving and exporting data 17 5.1 Saving data in an R format ...................... 17 5.2 Exporting data into a csv file ..................... 18 5.3 Exporting data with the openxlsx package ............. 19 6 Graphical data analysis with the ggplot2 package 20 6.1 Line plots ............................... 20 6.2 Bar charts ............................... 24 6.3 Scatterplots .............................. 25 6.4 Saving plots .............................. 27 7 Loops and random numbers 28 7.1 Cross-sectional data .......................... 28 7.2 Time series data ............................ 31 8 Estimating econometric models 33 8.1 Linear regression with cross sectional data .............. 34 8.2 ARIMA models ............................ 37 9 References 38 9.1 Literature ............................... 38 9.2 Further resources ........................... 38 1 2 INSTALLATION 1 Introduction Data analysis is becoming increasingly important in today’s professional life and learning some basic data science skills appears to be a worthwhile effort. It can boost your productivity at work and improve your attractiveness on the job market. Among the myriad of programming languages out there, R has emerged as one of the most popular starting points for such an endeavour. It is a powerful application to analyse data, it is free and has a steep learning curve. -

Datascientist Manual
DATASCIENTIST MANUAL . 2 « Approcherait le comportement de la réalité, celui qui aimerait s’epanouir dans l’holistique, l’intégratif et le multiniveaux, l’énactif, l’incarné et le situé, le probabiliste et le non linéaire, pris à la fois dans l’empirique, le théorique, le logique et le philosophique. » 4 (* = not yet mastered) THEORY OF La théorie des probabilités en mathématiques est l'étude des phénomènes caractérisés PROBABILITY par le hasard et l'incertitude. Elle consistue le socle des statistiques appliqué. Rubriques ↓ Back to top ↑_ Notations Formalisme de Kolmogorov Opération sur les ensembles Probabilités conditionnelles Espérences conditionnelles Densités & Fonctions de répartition Variables aleatoires Vecteurs aleatoires Lois de probabilités Convergences et théorèmes limites Divergences et dissimilarités entre les distributions Théorie générale de la mesure & Intégration ------------------------------------------------------------------------------------------------------------------------------------------ 6 Notations [pdf*] Formalisme de Kolmogorov Phé nomé né alé atoiré Expé riéncé alé atoiré L’univérs Ω Ré alisation éléméntairé (ω ∈ Ω) Evé némént Variablé aléatoiré → fonction dé l’univér Ω Opération sur les ensembles : Union / Intersection / Complémentaire …. Loi dé Augustus dé Morgan ? Independance & Probabilités conditionnelles (opération sur des ensembles) Espérences conditionnelles 8 Variables aleatoires (discret vs reel) … Vecteurs aleatoires : Multiplét dé variablés alé atoiré (discret vs reel) … Loi marginalés Loi -

Opening Reproducible Research: Research Project Website and Blog Blog Post Authors
Opening Reproducible Research: research project website and blog Blog post authors: Daniel Nüst Lukas Lohoff Marc Schutzeichel Markus Konkol Matthias Hinz Rémi Rampin Vicky Steeves Opening Reproducible Research | doi:10.5281/zenodo.1485438 Blog posts Demo server update 14 Aug 2018 | By Daniel Nüst We’ve been working on demonstrating our reference-implementation during spring an managed to create a number of example workspaces. We now decided to publish these workspaces on our demo server. Screenshot 1: o2r reference implementation listing of published Executable Research Compendia . The right-hand side shows a metadata summary including original authors. The papers were originally published in Journal of Statistical Software or in a Copernicus Publications journal under open licenses. We have created an R Markdown document for each paper based on the included data and code following the ERC specification for naming core files, but only included data, an R Markdown document and a HTML display file. The publication metadata, the runtime environment description (i.e. a Dockerfile ), and the runtime image (i.e. a Docker image tarball) were all created during the ERC creation process without any human interaction (see the used R code for upload), since required metadata were included in the R Markdown document’s front matter. The documents include selected figures or in some cases the whole paper, if runtime is not extremely long. While the paper’s authors are correctly linked in the workspace metadata (see right hand side in Screenshot 1), the “o2r author” of all papers is o2r team member Daniel since he made the uploads.