Imagej2: Imagej for the Next Generation of Scientific Image Data Curtis T
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Return of Organization Exempt from Income
OMB No. 1545-0047 Return of Organization Exempt From Income Tax Form 990 Under section 501(c), 527, or 4947(a)(1) of the Internal Revenue Code (except black lung benefit trust or private foundation) Open to Public Department of the Treasury Internal Revenue Service The organization may have to use a copy of this return to satisfy state reporting requirements. Inspection A For the 2011 calendar year, or tax year beginning 5/1/2011 , and ending 4/30/2012 B Check if applicable: C Name of organization The Apache Software Foundation D Employer identification number Address change Doing Business As 47-0825376 Name change Number and street (or P.O. box if mail is not delivered to street address) Room/suite E Telephone number Initial return 1901 Munsey Drive (909) 374-9776 Terminated City or town, state or country, and ZIP + 4 Amended return Forest Hill MD 21050-2747 G Gross receipts $ 554,439 Application pending F Name and address of principal officer: H(a) Is this a group return for affiliates? Yes X No Jim Jagielski 1901 Munsey Drive, Forest Hill, MD 21050-2747 H(b) Are all affiliates included? Yes No I Tax-exempt status: X 501(c)(3) 501(c) ( ) (insert no.) 4947(a)(1) or 527 If "No," attach a list. (see instructions) J Website: http://www.apache.org/ H(c) Group exemption number K Form of organization: X Corporation Trust Association Other L Year of formation: 1999 M State of legal domicile: MD Part I Summary 1 Briefly describe the organization's mission or most significant activities: to provide open source software to the public that we sponsor free of charge 2 Check this box if the organization discontinued its operations or disposed of more than 25% of its net assets. -
UMD) Open Source Software Packages
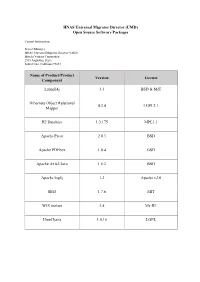
HNAS Universal Migrator Director (UMD) Open Source Software Packages Contact Information: Project Manager HNAS Universal Migrator Director (UMD) Hitachi Vantara Corporation 2535 Augustine Drive Santa Clara, California 95054 Name of Product/Product Version License Component Launch4j 3.3 BSD & MIT Hibernate Object Relational 4.3.4 LGPL2.1 Mapper H2 Database 1.3.175 MPL1.1 Apache Pivot 2.0.3 BSD Apache PDFbox 1.8.4 BSD Apache Axis2/Java 1.6.2 BSD Apache log4j 1.2 Apache v2.0 Slf4J 1.7.6 MIT WIX toolset 3.8 Ms-RL JfreeCharts 1.0.16 LGPL The text of the open source software licenses listed in this document is provided in the Open Source Software License Terms document on www.hitachivantara.com. Note that the source code for packages licensed under the GNU General Public License or similar type of license that requires the licensor to make the source code publicly available (“GPL Software”) may be available for download as indicated. If the source code for GPL Software is not included in the software or available for download, please send requests for source code for GPL Software to the contact person listed above for this product. The material in this document is provided “AS IS,” without warranty of any kind, including, but not limited to, the implied warranties of merchantability, fitness for a particular purpose, and non-infringement. Unless specified in an applicable open source license, access to this material grants you no right or license, express or implied, statutorily or otherwise, under any patent, trade secret, copyright, or any other intellectual property right of Hitachi Vantara Corporation (“Hitachi”). -

2. Enterprise Architecture 2.4 User Interaction Capabilities
VA Enterprise Design Patterns: 2. Enterprise Architecture 2.4 User Interaction Capabilities Office of Technology Strategies (TS) Architecture, Strategy, and Design (ASD) Office of Information and Technology (OI&T) Version 1.0 Date Issued: October 2015 THIS PAGE INTENTIONALLY LEFT BLANK FOR PRINTING PURPOSES APPROVAL COORDINATION Digitally signed by TIMOTHY L MCGRAIL 111224 TIMOTHY L DN: dc=gov, dc=va, o=internal, ou=people, MCGRAIL 0.9.2342.19200300.100.1.1=tim. [email protected], cn=TIMOTHY L MCGRAIL 111224 Date: 111224 Date: 2015.11.17 08:23:43 -05'00' Tim McGrail Senior Program Analyst ASD Technology Strategies Digitally signed by PAUL A. TIBBITS PAUL A. 116858 DN: dc=gov, dc=va, o=internal, ou=people, TIBBITS 0.9.2342.19200300.100.1.1=paul.tibbits@ va.gov, cn=PAUL A. TIBBITS 116858 Date: Reason: I am approving this document. 116858 Date: 2015.11.30 12:29:02 -05'00' Paul A. Tibbits, M.D. DCIO Architecture, Strategy, and Design REVISION HISTORY Version Date Organization Notes 0.1 2/27/2015 ASD TS Initial Draft Updated draft to reflect additional research and inputs 0.3 5/8/2015 ASD TS from vendor engagements and Stakeholders, with QA provided by TS DP Contractor Team Updated draft to reflect feedback from the Public 0.5 5/22/2015 ASD TS Forum held on 13 May 2015, and QA provided by ASD TS Updated version to align to the latest template for 0.7 10/13/15 ASD TS Enterprise Design Patterns and to account for internal QA review 0.9 REVISION HISTORY APPROVALS Version Date Approver Role 0.1 2/27/2015 Joseph Brooks ASD TS Design Pattern Government Lead 0.3 5/8/2015 Joseph Brooks ASD TS Design Pattern Government Lead 0.5 5/22/2015 Joseph Brooks ASD TS Design Pattern Government Lead 0.7 10/13/15 Joseph Brooks ASD TS Design Pattern Government Lead 1.0 11/17/15 Tim McGrail ASD TS Design Pattern Final Review TABLE OF CONTENTS 1.0 INTRODUCTION ........................................................................................................................................ -

The Power of Interoperability: Why Objects Are Inevitable
The Power of Interoperability: Why Objects Are Inevitable Jonathan Aldrich Carnegie Mellon University Pittsburgh, PA, USA [email protected] Abstract 1. Introduction Three years ago in this venue, Cook argued that in Object-oriented programming has been highly suc- their essence, objects are what Reynolds called proce- cessful in practice, and has arguably become the dom- dural data structures. His observation raises a natural inant programming paradigm for writing applications question: if procedural data structures are the essence software in industry. This success can be documented of objects, has this contributed to the empirical success in many ways. For example, of the top ten program- of objects, and if so, how? ming languages at the LangPop.com index, six are pri- This essay attempts to answer that question. After marily object-oriented, and an additional two (PHP reviewing Cook’s definition, I propose the term ser- and Perl) have object-oriented features.1 The equiva- vice abstractions to capture the essential nature of ob- lent numbers for the top ten languages in the TIOBE in- jects. This terminology emphasizes, following Kay, that dex are six and three.2 SourceForge’s most popular lan- objects are not primarily about representing and ma- guages are Java and C++;3 GitHub’s are JavaScript and nipulating data, but are more about providing ser- Ruby.4 Furthermore, objects’ influence is not limited vices in support of higher-level goals. Using examples to object-oriented languages; Cook [8] argues that Mi- taken from object-oriented frameworks, I illustrate the crosoft’s Component Object Model (COM), which has unique design leverage that service abstractions pro- a C language interface, is “one of the most pure object- vide: the ability to define abstractions that can be ex- oriented programming models yet defined.” Academ- tended, and whose extensions are interoperable in a ically, object-oriented programming is a primary focus first-class way. -

SWG1 N153 2010-08-26 Title: Open Source Software Category
SWG1 N153 2010-08-26 Title: Open Source Software Category Date Assigned: 2010-08-26 Source: Japan MB Backward Pointer: None Document Type: Status: This document will be discussed on First OMATF meeting. Open Source Software Category No Software Category No Software 1-1 OS Linux 1 Andoroid 2 Debian 3 Fedora 4 Google Chrome OS 5 openSUSE 6 Ubuntu 7 Vine Linux 1-2 BSD 1 FreeBSD 2 NetBSD 3 OpenBSD 1-3 Other 1 OpenSolaris 2 Symbian OS 2-1 Internet Server WEB 1 Apache HTTP Server 2 Appweb 3 Jetty 4 lighttpd 5 Mongoose 6 Open Web Server 7 TUX Web Server 2-2 SMTP:Simple Mail Transfer Protocol 1 Courier-MTA 2 Postfix 3 qmail 4 sendmail 5 XMail 2-3 POP:Post Office Protocol/IMAP:Internet Message Access Protocol 1 Courier IMAP 2 Cyrus IMAP 3 Dovecot 4 qpopper 2-4 Proxy 1 Squid Cache 2 Delegate 3 Tor 2-5 DNS:Domain Name System 1 BIND 2 djbdns 3 NSD 4 unbound 2-6 DHCP:Dynamic Host Configuration Protocol 1 DHCP 2-7 LDAP:Lightweight Directory Access Protocol 1 Apache Directory Server 2 Fedora Directory Server 3 OpenDS 4 OpenLDAP 2-8 Cooperation with Windows 1 Samba 2-9 FTP:File Transfer Protocol 1 FileZilla 2 ProFTPD 3 publicfile 4 Pure-FTPd 5 vsftpd 6 WU-FTPD 7 zFTPServer 3-1Application Server Java Platform, Enterprise Edition Server 1 Apache Geronimo 2 Apache Tomcat 3 Enhydra Server 4 GlassFish 5 JBoss Application Server 6 JOnAS 7 Seasar2 3-2 Other Java Platform, Enterprise Edition Server 1 Seasar2 2 Zope 3-3 Execution Environment 1 GCC 2 OpenJDK 3-4 Script 1 Perl 2 PHP 3 Python 4 Ruby 4-1Database DBMS: DataBase Management System 1 Apache Derby 2 Firebird -

Thesis Entire.Pdf (12.90Mb)
Faculty OF Science AND TECHNOLOGY Department OF Computer Science TOWARD ReprODUCIBLE Analysis AND ExplorATION OF High-ThrOUGHPUT Biological Datasets — Bjørn Fjukstad A DISSERTATION FOR THE DEGREE OF Philosophiae Doctor – 2018 This thesis document was typeset using the UiT Thesis LaTEX Template. © 2018 – http://github.com/egraff/uit-thesis “Ta aldri problemene på forskudd, for da får du dem to ganger, men ta gjerne seieren på forskudd, for hvis ikke er det altfor sjelden du får oppleve den.” –Ivar Tollefsen AbstrACT There is a rapid growth in the number of available biological datasets due to the advent of high-throughput data collection instruments combined with cheap compute infrastructure. Modern instruments enable the analysis of biological data at different levels, from small DNA sequences through larger cell structures, and up to the function of entire organs. These new datasets have brought the need to develop new software packages to enable novel insights into the underlying biological mechanisms in the development and progression of diseases such as cancer. The heterogeneity of biological datasets require researchers to tailor the explo- ration and analyses with a wide range of different tools and systems. However, despite the need for their integration, few of them provide standard inter- faces for analyses implemented using different programming languages and frameworks. In addition, because of the many tools, different input parame- ters, and references to databases, it is necessary to record these correctly. The lack of such details complicates reproducing the original results and the reuse of the analyses on new datasets. This increases the analysis time and leaves unrealized potential for scientific insights. -

AVR Simulator
AVR Simulator Ryan Moore - Colorado State University February 8, 2011 1 Overview The AVR Simulator is designed to simulate the minimal number of instructions that is required to program the MeggyJr RGB device. The simulator can either run in a batch mode, where the assembly program will run to completion or till a system imposed hop count occurs, or in a gui mode where there is visualization for the stack, heap, registers and program space (there is a planned update to include a visualization of the actual device with button presses). The simulator is entirely written in Java and uses several open source api's. The simulator is distributed as an executable jar and does not require any other les/libraries in order to run. Using the Simulator In order to use the simulator you must invoke the java jvm with the jar option: java -jar MJSIM.jar This will invoke the java jvm and execute the simulator in gui mode. The default behavior is to load the gui, and then a user can open an assembly le within the application, and then step through, or run, the assembly program. The default behavior can be modied with dierent command line options. The most important of these is the batch mode option and the le option. To invoke the simulator in batch mode add the -b option after MJSIM.jar: java -jar MJSIM.jar -b This command will invoke the simulator in batch mode. This however will not run. When executing the simulator in batch mode a le is required as well. -

JSR 377 Desktop Application Framework May 24Th 2018 Andres Almiray Agenda
JSR 377 Desktop Application Framework May 24th 2018 Andres Almiray Agenda • Goals • Information to be gathered • Implementation notes • Issues • Questions, discussion, next steps 2 Goals 3 Goals • Define APIs that can be used to build Java Desktop (and possible mobile) applications. • Inspired in previous efforts such as JSR 296 (Swing Application Framework) and JSR 295 (Beans Binding) • JSR 377 does not target a single UI toolkit. It should be possible to use existing toolkits such as Swing, JavaFX, SWT, Apache Pivot, Lanterna, others. 4 Information to be gathered 5 About this JSR • Provide the following features application life cycle localized resources (and injection) persisted session state loosely coupled actions dependency injection event system centralized error management extension points via plugins 6 Introduction • JSR 193 – Client Side Container • JSR 296 – Swing Application Framework • JSR 295 – Beans Binding • JSR 296 had the following goals application life cycle localized resources (and injection) persisted session state loosely coupled actions targeted only Swing for obvious reasons 7 Business/marketing/ecosystem justification • Why do this JSR? – Desktop application development can benefit form lessons learned by JavaEE standardization. • What's the need? – Applications can’t share code/components unless they are written using the same base platform (NetBeans, RCP, Eclipse RCP, homegrown). • How does it fit in to the Java ecosystem? – Desktop applications are still being built. • Is the idea ready for standardization? – UI toolkits and desktop platforms are mature enough. 8 History • List the significant dates in the history of the JSR. – Submitted on 29 Dec, 2014. – Review Ballots: 27 Jan, 2015 16 Feb, 2016 14 Mar, 2017 – API drafted. -

Download Final Mlearn Con Flash and HTML5. for Distrib
Why I Stand With (Adobe) Flash Ellen Wagner Sage Road Solutions LLC June 15, 2010 © 2010 Adobe Systems Incorporated. All rights reserved. Used With permission, mLearnCon, June 15, 2010 Adobe, the Adobe logo, Adobe AIR, the Adobe AIR logo, the Adobe PDF logo, AIR, ColdFusion, ColdFusion Builder, Flash, Flash Builder, the Flash logo, Flex, LiveCycle, and Reader are either registered trademarks or trademarks of Adobe Systems Incorporated in the United States and/or other countries. Do You Know What Flash Is? “It's a serious question. During the past few months, volumes and volumes have been written about Flash. It's dead. It's dying. It's lazy, buggy and bloated. It's everywhere. It's efficient, it makes the web work. It's ubiquitous. It creates rich, engaging digital experience. It is going to drive the knowledge commerce economy. It's proprietary. It’s mostly open. It is irrelevant. In its future iterations, it's where the mobile Web is headed.” http://elearningroadtrip.typepad.com/elearning_roadtrip/2010/05/flash- primer-for-non-techies.html The Many Faces of Flash • It's a file format. Most commonly known as SWF. Like PDF for web content. • It's a free media player available on the web. You need it to view SWF files - movies, video, animations. Comes in two forms: Flash Player and AIR. • It provides a way to share animations, interactive content and video on the web. • It's a web animation scripting software, known by the name of Flash Professional. • It provides a Flash-based path for creating Rich Internet Applications (RIAs) • It's a published web specification. -

Code Smell Prediction Employing Machine Learning Meets Emerging Java Language Constructs"
Appendix to the paper "Code smell prediction employing machine learning meets emerging Java language constructs" Hanna Grodzicka, Michał Kawa, Zofia Łakomiak, Arkadiusz Ziobrowski, Lech Madeyski (B) The Appendix includes two tables containing the dataset used in the paper "Code smell prediction employing machine learning meets emerging Java lan- guage constructs". The first table contains information about 792 projects selected for R package reproducer [Madeyski and Kitchenham(2019)]. Projects were the base dataset for cre- ating the dataset used in the study (Table I). The second table contains information about 281 projects filtered by Java version from build tool Maven (Table II) which were directly used in the paper. TABLE I: Base projects used to create the new dataset # Orgasation Project name GitHub link Commit hash Build tool Java version 1 adobe aem-core-wcm- www.github.com/adobe/ 1d1f1d70844c9e07cd694f028e87f85d926aba94 other or lack of unknown components aem-core-wcm-components 2 adobe S3Mock www.github.com/adobe/ 5aa299c2b6d0f0fd00f8d03fda560502270afb82 MAVEN 8 S3Mock 3 alexa alexa-skills- www.github.com/alexa/ bf1e9ccc50d1f3f8408f887f70197ee288fd4bd9 MAVEN 8 kit-sdk-for- alexa-skills-kit-sdk- java for-java 4 alibaba ARouter www.github.com/alibaba/ 93b328569bbdbf75e4aa87f0ecf48c69600591b2 GRADLE unknown ARouter 5 alibaba atlas www.github.com/alibaba/ e8c7b3f1ff14b2a1df64321c6992b796cae7d732 GRADLE unknown atlas 6 alibaba canal www.github.com/alibaba/ 08167c95c767fd3c9879584c0230820a8476a7a7 MAVEN 7 canal 7 alibaba cobar www.github.com/alibaba/ -

Opening Reproducible Research: Research Project Website and Blog Blog Post Authors
Opening Reproducible Research: research project website and blog Blog post authors: Daniel Nüst Lukas Lohoff Marc Schutzeichel Markus Konkol Matthias Hinz Rémi Rampin Vicky Steeves Opening Reproducible Research | doi:10.5281/zenodo.1485438 Blog posts Demo server update 14 Aug 2018 | By Daniel Nüst We’ve been working on demonstrating our reference-implementation during spring an managed to create a number of example workspaces. We now decided to publish these workspaces on our demo server. Screenshot 1: o2r reference implementation listing of published Executable Research Compendia . The right-hand side shows a metadata summary including original authors. The papers were originally published in Journal of Statistical Software or in a Copernicus Publications journal under open licenses. We have created an R Markdown document for each paper based on the included data and code following the ERC specification for naming core files, but only included data, an R Markdown document and a HTML display file. The publication metadata, the runtime environment description (i.e. a Dockerfile ), and the runtime image (i.e. a Docker image tarball) were all created during the ERC creation process without any human interaction (see the used R code for upload), since required metadata were included in the R Markdown document’s front matter. The documents include selected figures or in some cases the whole paper, if runtime is not extremely long. While the paper’s authors are correctly linked in the workspace metadata (see right hand side in Screenshot 1), the “o2r author” of all papers is o2r team member Daniel since he made the uploads. -

Jsr223: a Java Platform Integration for R with Programming Languages Groovy, Javascript, Jruby, Jython, and Kotlin by Floid R
CONTRIBUTED RESEARCH ARTICLES 440 jsr223: A Java Platform Integration for R with Programming Languages Groovy, JavaScript, JRuby, Jython, and Kotlin by Floid R. Gilbert and David B. Dahl Abstract The R package jsr223 is a high-level integration for five programming languages in the Java platform: Groovy, JavaScript, JRuby, Jython, and Kotlin. Each of these languages can use Java objects in their own syntax. Hence, jsr223 is also an integration for R and the Java platform. It enables developers to leverage Java solutions from within R by embedding code snippets or evaluating script files. This approach is generally easier than rJava’s low-level approach that employs the Java Native Interface. jsr223’s multi-language support is dependent on the Java Scripting API: an implementation of “JSR-223: Scripting for the Java Platform” that defines a framework to embed scripts in Java applications. The jsr223 package also features extensive data exchange capabilities and a callback interface that allows embedded scripts to access the current R session. In all, jsr223 makes solutions developed in Java or any of the jsr223-supported languages easier to use in R. Introduction About the same time Ross Ihaka and Robert Gentleman began developing R at the University of Auckland in the early 1990s, James Gosling and the so-called Green Project Team was working on a new programming language at Sun Microsystems in California. The Green Team did not set out to make a new language; rather, they were trying to move platform-independent, distributed computing into the consumer electronics marketplace. As Gosling explained, “All along, the language was a tool, not the end” (O’Connell, 1995).