Species' Traits and Phylogenetic
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Special Publications Museum of Texas Tech University Number 63 18 September 2014
Special Publications Museum of Texas Tech University Number 63 18 September 2014 List of Recent Land Mammals of Mexico, 2014 José Ramírez-Pulido, Noé González-Ruiz, Alfred L. Gardner, and Joaquín Arroyo-Cabrales.0 Front cover: Image of the cover of Nova Plantarvm, Animalivm et Mineralivm Mexicanorvm Historia, by Francisci Hernández et al. (1651), which included the first list of the mammals found in Mexico. Cover image courtesy of the John Carter Brown Library at Brown University. SPECIAL PUBLICATIONS Museum of Texas Tech University Number 63 List of Recent Land Mammals of Mexico, 2014 JOSÉ RAMÍREZ-PULIDO, NOÉ GONZÁLEZ-RUIZ, ALFRED L. GARDNER, AND JOAQUÍN ARROYO-CABRALES Layout and Design: Lisa Bradley Cover Design: Image courtesy of the John Carter Brown Library at Brown University Production Editor: Lisa Bradley Copyright 2014, Museum of Texas Tech University This publication is available free of charge in PDF format from the website of the Natural Sciences Research Laboratory, Museum of Texas Tech University (nsrl.ttu.edu). The authors and the Museum of Texas Tech University hereby grant permission to interested parties to download or print this publication for personal or educational (not for profit) use. Re-publication of any part of this paper in other works is not permitted without prior written permission of the Museum of Texas Tech University. This book was set in Times New Roman and printed on acid-free paper that meets the guidelines for per- manence and durability of the Committee on Production Guidelines for Book Longevity of the Council on Library Resources. Printed: 18 September 2014 Library of Congress Cataloging-in-Publication Data Special Publications of the Museum of Texas Tech University, Number 63 Series Editor: Robert J. -

Sweave User Manual
Sweave User Manual Friedrich Leisch and R-core June 30, 2017 1 Introduction Sweave provides a flexible framework for mixing text and R code for automatic document generation. A single source file contains both documentation text and R code, which are then woven into a final document containing • the documentation text together with • the R code and/or • the output of the code (text, graphs) This allows the re-generation of a report if the input data change and documents the code to reproduce the analysis in the same file that contains the report. The R code of the complete 1 analysis is embedded into a LATEX document using the noweb syntax (Ramsey, 1998) which is usually used for literate programming Knuth(1984). Hence, the full power of LATEX (for high- quality typesetting) and R (for data analysis) can be used simultaneously. See Leisch(2002) and references therein for more general thoughts on dynamic report generation and pointers to other systems. Sweave uses a modular concept using different drivers for the actual translations. Obviously different drivers are needed for different text markup languages (LATEX, HTML, . ). Several packages on CRAN provide support for other word processing systems (see Appendix A). 2 Noweb files noweb (Ramsey, 1998) is a simple literate-programming tool which allows combining program source code and the corresponding documentation into a single file. A noweb file is a simple text file which consists of a sequence of code and documentation segments, called chunks: Documentation chunks start with a line that has an at sign (@) as first character, followed by a space or newline character. -

45762554013.Pdf
Mastozoología Neotropical ISSN: 0327-9383 ISSN: 1666-0536 [email protected] Sociedad Argentina para el Estudio de los Mamíferos Argentina Flores-Alta, Daniel; Rivera-Ortíz, Francisco A.; Contreras-González, Ana M. RECORD OF A POPULATION AND DESCRIPTION OF SOME ASPECTS OF THE LIFE HISTORY OF Notocitellus adocetus IN THE NORTH OF THE STATE OF GUERRERO, MEXICO Mastozoología Neotropical, vol. 26, no. 1, 2019, -June, pp. 175-181 Sociedad Argentina para el Estudio de los Mamíferos Argentina Available in: https://www.redalyc.org/articulo.oa?id=45762554013 How to cite Complete issue Scientific Information System Redalyc More information about this article Network of Scientific Journals from Latin America and the Caribbean, Spain and Journal's webpage in redalyc.org Portugal Project academic non-profit, developed under the open access initiative Mastozoología Neotropical, 26(1):175-181, Mendoza, 2019 Copyright ©SAREM, 2019 Versión on-line ISSN 1666-0536 http://www.sarem.org.ar https://doi.org/10.31687/saremMN.19.26.1.0.02 http://www.sbmz.com.br Nota RECORD OF A POPULATION AND DESCRIPTION OF SOME ASPECTS OF THE LIFE HISTORY OF Notocitellus adocetus IN THE NORTH OF THE STATE OF GUERRERO, MEXICO Daniel Flores-Alta, Francisco A. Rivera-Ortíz and Ana M. Contreras-González Unidad de Biotecnología y Prototipos, Laboratorio de Ecología, Facultad de Estudios Superiores Iztacala, Universidad Nacional Autónoma de México, Los Reyes Iztacala. Tlalnepantla, Estado de México, México [correspondencia: Ana M. Contreras-González <[email protected]>]. ABSTRACT. The rodent Notocitellus adocetus is endemic to Mexico, and little is known about its life history. We describe a population of N. -

Controlled Animals
Environment and Sustainable Resource Development Fish and Wildlife Policy Division Controlled Animals Wildlife Regulation, Schedule 5, Part 1-4: Controlled Animals Subject to the Wildlife Act, a person must not be in possession of a wildlife or controlled animal unless authorized by a permit to do so, the animal was lawfully acquired, was lawfully exported from a jurisdiction outside of Alberta and was lawfully imported into Alberta. NOTES: 1 Animals listed in this Schedule, as a general rule, are described in the left hand column by reference to common or descriptive names and in the right hand column by reference to scientific names. But, in the event of any conflict as to the kind of animals that are listed, a scientific name in the right hand column prevails over the corresponding common or descriptive name in the left hand column. 2 Also included in this Schedule is any animal that is the hybrid offspring resulting from the crossing, whether before or after the commencement of this Schedule, of 2 animals at least one of which is or was an animal of a kind that is a controlled animal by virtue of this Schedule. 3 This Schedule excludes all wildlife animals, and therefore if a wildlife animal would, but for this Note, be included in this Schedule, it is hereby excluded from being a controlled animal. Part 1 Mammals (Class Mammalia) 1. AMERICAN OPOSSUMS (Family Didelphidae) Virginia Opossum Didelphis virginiana 2. SHREWS (Family Soricidae) Long-tailed Shrews Genus Sorex Arboreal Brown-toothed Shrew Episoriculus macrurus North American Least Shrew Cryptotis parva Old World Water Shrews Genus Neomys Ussuri White-toothed Shrew Crocidura lasiura Greater White-toothed Shrew Crocidura russula Siberian Shrew Crocidura sibirica Piebald Shrew Diplomesodon pulchellum 3. -

ESS — Emacs Speaks Statistics
ESS — Emacs Speaks Statistics ESS version 5.14 The ESS Developers (A.J. Rossini, R.M. Heiberger, K. Hornik, M. Maechler, R.A. Sparapani, S.J. Eglen, S.P. Luque and H. Redestig) Current Documentation by The ESS Developers Copyright c 2002–2010 The ESS Developers Copyright c 1996–2001 A.J. Rossini Original Documentation by David M. Smith Copyright c 1992–1995 David M. Smith Permission is granted to make and distribute verbatim copies of this manual provided the copyright notice and this permission notice are preserved on all copies. Permission is granted to copy and distribute modified versions of this manual under the conditions for verbatim copying, provided that the entire resulting derived work is distributed under the terms of a permission notice identical to this one. Chapter 1: Introduction to ESS 1 1 Introduction to ESS The S family (S, Splus and R) and SAS statistical analysis packages provide sophisticated statistical and graphical routines for manipulating data. Emacs Speaks Statistics (ESS) is based on the merger of two pre-cursors, S-mode and SAS-mode, which provided support for the S family and SAS respectively. Later on, Stata-mode was also incorporated. ESS provides a common, generic, and useful interface, through emacs, to many statistical packages. It currently supports the S family, SAS, BUGS/JAGS, Stata and XLisp-Stat with the level of support roughly in that order. A bit of notation before we begin. emacs refers to both GNU Emacs by the Free Software Foundation, as well as XEmacs by the XEmacs Project. The emacs major mode ESS[language], where language can take values such as S, SAS, or XLS. -

Body Size and Diet Mediate Evolution of Jaw Shape in Squirrels (Sciuridae)
Rare ecomorphological convergence on a complex adaptive landscape: body size and diet mediate evolution of jaw shape in squirrels (Sciuridae) *Miriam Leah Zelditch; Museum of Paleontology, University of Michigan, Ann Arbor, MI 48109; [email protected] Ji Ye; Earth and Environmental Sciences, University of Michigan, Ann Arbor, MI 48109; [email protected] Jonathan S. Mitchell; Ecology and Evolutionary Biology, University of Michigan, Ann Arbor, MI 48109; [email protected] Donald L. Swiderski, Kresge Hearing Research Institute and Museum of Zoology, University of Michigan. [email protected] *Corresponding Author Running head: Convergence on a complex adaptive landscape Key Words: Convergence, diet evolution, jaw morphology, shape evolution, macroevolutionary adaptive landscape, geometric morphometrics Data archival information: doi: 10.5061/dryad.kq1g6. This is the author manuscript accepted for publication and has undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: 10.1111/evo.13168. This article is protected by copyright. All rights reserved. Abstract Convergence is widely regarded as compelling evidence for adaptation, often being portrayed as evidence that phenotypic outcomes are predictable from ecology, overriding contingencies of history. However, repeated outcomes may be very rare unless adaptive landscapes are simple, structured by strong ecological and functional constraints. One such constraint may be a limitation on body size because performance often scales with size, allowing species to adapt to challenging functions by modifying only size. When size is constrained, species might adapt by changing shape; convergent shapes may therefore be common when size is limiting and functions are challenging. -

How Many Species of Mammals Are There?
Journal of Mammalogy, 99(1):1–14, 2018 DOI:10.1093/jmammal/gyx147 INVITED PAPER How many species of mammals are there? CONNOR J. BURGIN,1 JOCELYN P. COLELLA,1 PHILIP L. KAHN, AND NATHAN S. UPHAM* Department of Biological Sciences, Boise State University, 1910 University Drive, Boise, ID 83725, USA (CJB) Department of Biology and Museum of Southwestern Biology, University of New Mexico, MSC03-2020, Albuquerque, NM 87131, USA (JPC) Museum of Vertebrate Zoology, University of California, Berkeley, CA 94720, USA (PLK) Department of Ecology and Evolutionary Biology, Yale University, New Haven, CT 06511, USA (NSU) Integrative Research Center, Field Museum of Natural History, Chicago, IL 60605, USA (NSU) 1Co-first authors. * Correspondent: [email protected] Accurate taxonomy is central to the study of biological diversity, as it provides the needed evolutionary framework for taxon sampling and interpreting results. While the number of recognized species in the class Mammalia has increased through time, tabulation of those increases has relied on the sporadic release of revisionary compendia like the Mammal Species of the World (MSW) series. Here, we present the Mammal Diversity Database (MDD), a digital, publically accessible, and updateable list of all mammalian species, now available online: https://mammaldiversity.org. The MDD will continue to be updated as manuscripts describing new species and higher taxonomic changes are released. Starting from the baseline of the 3rd edition of MSW (MSW3), we performed a review of taxonomic changes published since 2004 and digitally linked species names to their original descriptions and subsequent revisionary articles in an interactive, hierarchical database. We found 6,495 species of currently recognized mammals (96 recently extinct, 6,399 extant), compared to 5,416 in MSW3 (75 extinct, 5,341 extant)—an increase of 1,079 species in about 13 years, including 11 species newly described as having gone extinct in the last 500 years. -

Reproducible Research.. Using Sweave, Knitr and Pandoc
Reproducible Research.. Using Sweave, Knitr and Pandoc Aedín Culhane [email protected] Nov 20th 2012 My R Course Website http://bcb.dfci.harvard.edu/~aedin/ My HSPH homepage http://www.hsph.harvard.edu/research/aedin-culhane/ When issues of reproducibility arise • ``Remember that microarray analysis you did six months ago? We ran a few more arrays. Can you add them to the project and repeat the same analysis?'' • ``The statistical analyst who looked at the data I generated previously is no longer available. Can you get someone else to analyze my new data set using the same methods (and thus producing a report I can expect to understand)?'' • ``Please write/edit the methods sections for the abstract/paper/grant proposal I am submitting based on the analysis you did several months ago.'' From Keith Baggerly • Selected articles published in Nature Genetics between January 2005 and December 2006 that had used profiling with microarrays • Of the 56 items retrieved electronically, 20 articles were considered potentially eligible for the project • The four teams were from – University of Alabama at Birmingham (UAB) – Stanford/Dana-Farber (SD) – London (L) and Ioannina/Trento (IT) • Each team was comprised of 3-6 scientists who worked together to evaluate each article. Results • Result could be reproduced n=2 • Reproduced with discrepancy n=6 • Could not be reproduced n=10 – No data n=4 (no data n=2, subset n=1, no reporter data n=1) – Confusion over matching of data to analysis (n=2) – Specialized software required and not available (n=1)m – Raw data available but could not be processed n=2 Reproducibility of Analysis Ioannidis JP, Allison DB, Ball CA, Coulibaly I, Cui X, Culhane AC, et al, (2009) Repeatability of published microarray gene expression Analyses. -
Dynamic Documents with R and Knitr, by Yihui Xie 116 Tugboat, Volume 35 (2014), No
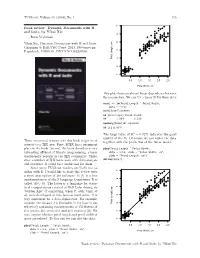
TUGboat, Volume 35 (2014), No. 1 115 ● ● ● Book review: Dynamic Documents with R ● ● ● and knitr, by Yihui Xie ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● Boris Veytsman ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● Yihui Xie, Dynamic Documents with R and knitr. ● ● ● ● ● ● ● ● ● ● ● ● ● ● Chapman & Hall/CRC Press, 2013, 190+xxvi pp. ● ● ● ● ● ● ● ● ● US$ ISBN ● Paperback, 59.95. 978-1482203530. ● ● ● ● Petal Length, cm Petal ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● 1 2 3 4 5 6 7 0.5 1.0 1.5 2.0 2.5 Petal Width, cm This plot shows an almost linear dependence between the parameters. We can try a linear fit for these data: model <- lm(Petal.Length ˜ Petal.Width, data = iris) model$coefficients ## (Intercept) Petal.Width ## 1.084 2.230 summary(model)$r.squared ## [1] 0.9271 The large value of R2 = 0.9271 indicates the good quality of the fit. Of course we can replot the data There are several reasons why this book might be of together with the prediction of the linear model: interest to a TEX user. First, LATEX has a prominent place in the book. Second, the book describes a very plot(Petal.Length ˜ Petal.Width, interesting offshoot of literate programming, a topic data = iris, xlab = "Petal Width, cm", traditionally popular in the TEX community. Third, ylab = "Petal Length, cm") abline(model) since a number of TEX users work with data analysis and statistics, R could be a useful tool for them. Since some TUGboat readers are likely not fa- ● ● ● miliar with R, I would like to start this review with ● ● a short description of the software. R [1] is a free ● ● ● ● ● ● ● ● ● ● ● implementation of the S language (sometimes R is ● ● ● ● ● ● ● ● ● ● ● ● ● ● called GNU S). -

LYX and Knitr
knitr.1 LYX and knitr The knitr package allows one to embed R code within LYX and LATEX documents. When a document is compiled into a PDF, LYX/LATEX connects to R to run the R code and the code/output is automatically put into the PDF. In addition to this being a convenient way to use both LYX/LATEX with R, it also provides an important component to the reproducibility of research (RR). For example, one can include the code for a data analysis de- scribed in a paper. This ensures that there would be no “copying and pasting errors” and also provide readers of the paper an im- mediate way to reproduce the research. RR continues to become more important and fortunately more tools are being developed to make it possible. Below are some discussions on the topic: • AMSTAT News column on RR at http://magazine. amstat.org/blog/2011/01/01/scipolicyjan11. • CRAN task view for RR and R at http://cran.r-project. org/web/views/ReproducibleResearch.html. • Yihui Xie: Author of knitr – First and second editions of his Dynamic Documents with R and knitr book. Note that this book was typed in LYX. – Website for knitr at http://yihui.name/knitr The Sweave environment is another way to include R code inside of LYX/LATEX. This was developed prior to knitr, but it is more difficult to use. The purpose of this section is to examine the main compo- nents of knitr so that you will be able to complete the rest of the semester using LYX and knitr together for all assign- ments in our course! Also, a very important purpose is to give you the tools needed to complete all assignments in other R- based courses by using knitr and LYX together! The files used knitr.2 here are intro_example_cereal.lyx, intro_example_cereal.pdf, cereal.csv, ExternalCode.R, FirstBeamer-knitr.zip, JSM2015.zip, and RMarkdown.zip. -

IRBP) in Ground Squirrels and Blind Mole Rats
Molecular Evolution of the Interphotoreceptor Retinoid Binding Protein (IRBP) in Ground Squirrels and Blind Mole Rats Research Thesis Presented in partial fulfillment of the requirements for graduation with Research Distinction in Biology in the Undergraduate Colleges of The Ohio State University by Bethany N. Army The Ohio State University December 2015 Project Advisor: Dr. Ryan W. Norris, Department of Evolution, Ecology and Organismal Biology Abstract Blind mole rats are subterranean rodents and spend most of their lifetime underground. Because these mammals live in a dark environment, their eyes have physically changed and even the role played by their eyes has changed. The processing of visual information begins in the retina, with the detection of light by photoreceptor cells. Interphotoreceptor retinoid binding protein (IRBP) is a transport protein, which facilitates exchange between photoreceptors and retinal-pigmented epithelium. The IRBP gene has been commonly used in rodent phylogenetic analyses, so I analyzed IRBP in blind mole rats to interpret its evolution and to see if there were any changes among the amino acids in the protein. To do this I used molecular data (cytochrome b and IRBP) from two groups of rodents, the muroids and the marmotines. I constructed phylogenetic trees and interpreted the amino acid sequence of IRBP. Blind mole rats had a slower rate of evolution than expected, but there were changes along the amino acid sequence of IRBP, which may indicate that the IRBP gene is functioning in a specialized way to meet the needs of the particular lifestyle of this mammal. Introduction: Blind mole rats, subfamily Spalacinae, are burrowing mammals that spend most of their life underground. -

Book of Abstracts
Book of Abstracts June 27, 2015 1 Conference Sponsors Diamond Sponsor Platinum Sponsors Gold Sponsors Silver Sponsors Open Analytics Bronze Sponsors Media Sponsors 2 Conference program Time Tuesday Wednesday Thursday Friday 08:00 Registration opens Registration opens Registration opens Registration opens 08:30 – 09:00 Opening session (by Rector peR! M. Johansen, Aalborg University) Aalborghallen 09:00 – 10:00 Romain François Di Cook Thomas Lumley Aalborghallen Aalborghallen Aalborghallen 10:00 – 10:30 Coffee break Coffee break Coffee break (15 min) ee break Sponsored by Quantide Sponsored by Alteryx ff Session 1 Session 4 10:30 – 12:00 Sponsor session (10:15) Kaleidoscope 1 Kaleidoscope 4 Aalborghallen Aalborghallen Aalborghallen incl. co Morning Tutorials DataRobot Ecology Medicine Gæstesalen Gæstesalen RStudio Teradata Networks Regression Musiksalen Musiksalen Revolution Analytics Reproducibility Commercial Offerings alteryx Det Lille Teater Det Lille Teater TIBCO H O Interfacing Interactive graphics 2 Radiosalen Radiosalen HP 12:00 – 13:00 Sandwiches Lunch (standing buffet) Lunch (standing buffet) Break: 12:00 – 12:30 Sponsored by Sponsored by TIBCO ff Revolution Analytics Ste en Lauritzen (12:30) Aalborghallen Session 2 Session 5 13:00 – 14:30 13:30: Closing remarks Kaleidoscope 2 Kaleidoscope 5 Aalborghallen Aalborghallen 13:45: Grab ’n go lunch 14:00: Conference ends Case study Teaching 1 Gæstesalen Gæstesalen Clustering Statistical Methodology 1 Musiksalen Musiksalen ee break Data Management Machine Learning 1 ff Det Lille Teater Det Lille