Durham Research Online
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Beaver's Phylogenetic Lineage Illuminated by Retroposon Reads
www.nature.com/scientificreports OPEN The Beaver’s Phylogenetic Lineage Illuminated by Retroposon Reads Liliya Doronina1,*, Andreas Matzke1,*, Gennady Churakov1,2, Monika Stoll3, Andreas Huge3 & Jürgen Schmitz1 Received: 13 October 2016 Solving problematic phylogenetic relationships often requires high quality genome data. However, Accepted: 25 January 2017 for many organisms such data are still not available. Among rodents, the phylogenetic position of the Published: 03 March 2017 beaver has always attracted special interest. The arrangement of the beaver’s masseter (jaw-closer) muscle once suggested a strong affinity to some sciurid rodents (e.g., squirrels), placing them in the Sciuromorpha suborder. Modern molecular data, however, suggested a closer relationship of beaver to the representatives of the mouse-related clade, but significant data from virtually homoplasy- free markers (for example retroposon insertions) for the exact position of the beaver have not been available. We derived a gross genome assembly from deposited genomic Illumina paired-end reads and extracted thousands of potential phylogenetically informative retroposon markers using the new bioinformatics coordinate extractor fastCOEX, enabling us to evaluate different hypotheses for the phylogenetic position of the beaver. Comparative results provided significant support for a clear relationship between beavers (Castoridae) and kangaroo rat-related species (Geomyoidea) (p < 0.0015, six markers, no conflicting data) within a significantly supported mouse-related clade (including Myodonta, Anomaluromorpha, and Castorimorpha) (p < 0.0015, six markers, no conflicting data). Most of an organism’s phylogenetic history is fossilized in their heritable genomic material. Using data from genome sequencing projects, particularly informative regions of this material can be extracted in sufficient num- bers to resolve the deepest history of speciation. -

Multiple Molecular Evidences for a Living Mammalian Fossil
Multiple molecular evidences for a living mammalian fossil Dorothe´ e Huchon†‡, Pascale Chevret§¶, Ursula Jordanʈ, C. William Kilpatrick††, Vincent Ranwez§, Paulina D. Jenkins‡‡, Ju¨ rgen Brosiusʈ, and Ju¨ rgen Schmitz‡ʈ †Department of Zoology, George S. Wise Faculty of Life Sciences, Tel Aviv University, Tel Aviv 69978, Israel; §Department of Paleontology, Phylogeny, and Paleobiology, Institut des Sciences de l’Evolution, cc064, Universite´Montpellier II, Place E. Bataillon, 34095 Montpellier Cedex 5, France; ʈInstitute of Experimental Pathology, University of Mu¨nster, D-48149 Mu¨nster, Germany; ††Department of Biology, University of Vermont, Burlington, VT 05405-0086; and ‡‡Department of Zoology, The Natural History Museum, London SW7 5BD, United Kingdom Edited by Francisco J. Ayala, University of California, Irvine, CA, and approved March 18, 2007 (received for review February 11, 2007) Laonastes aenigmamus is an enigmatic rodent first described in their classification as a diatomyid suggests that Laonastes is a 2005. Molecular and morphological data suggested that it is the living fossil and a ‘‘Lazarus taxon.’’ sole representative of a new mammalian family, the Laonastidae, The two research teams also disagreed on the taxonomic and a member of the Hystricognathi. However, the validity of this position of Laonastes. According to Jenkins et al. (2), Laonastes family is controversial because fossil-based phylogenetic analyses is either the most basal group of the hystricognaths (Fig. 2A)or suggest that Laonastes is a surviving member of the Diatomyidae, nested within the hystricognaths (Fig. 2B). According to Dawson a family considered to have been extinct for 11 million years. et al. (3), Laonastes and the other Diatomyidae are the sister According to these data, Laonastes and Diatomyidae are the sister clade of the family Ctenodactylidae (i.e., gundies), a family that clade of extant Ctenodactylidae (i.e., gundies) and do not belong does not belong to the Hystricognathi, but to which it is to the Hystricognathi. -

Chromosomal Numbers in African Giant Rat (Cricetomysgambianus, Waterhouse-1840)
IOSR Journal of Dental and Medical Sciences (IOSR-JDMS) e-ISSN: 2279-0853, p-ISSN: 2279-0861.Volume 18, Issue 7 Ser. 6 (July. 2019), PP 26-31 www.iosrjournals.org Chromosomal Numbers in African Giant Rat (Cricetomysgambianus, Waterhouse-1840) Ahmad, Im1, Musa, Sa2, Nzalak, Jo3 1Department of Anatomy, Faculty of Basic Medical Sciences, College of Health Sciences, UsmanuDanfodiyo University, Sokoto, Nigeria. 2Department of Human Anatomy, Faculty of Basic Medical Sciences, College of Health Sciences, Ahmadu Bello University, Zaria, Kaduna, Nigeria. 3Department of Veterinary Anatomy, Faculty of Veterinary Medicine, Ahmadu Bello University, Zaria. Kaduna, Nigeria. Corresponding Author: Ahmad, IM Abstract: Background: Karyotypic studies were carried out on the African giant rat, Cricetomysgambianus, Waterhouse-1840 with the aim of determining its chromosome diploid numbers and autosomal fundamental numbers. Methods: The chromosomes were prepared from the conventional bone marrow of two (2) African giant rats – a male and a female treated intra-peritoneally with 2 ml of 0.04% colchicines for 3 hours. Chromosomes in well spread mitotic metaphase cells were counted and measured using KaryoType computer software. Chromosomal numbers were identified. Ideograms were also constructed from the measurements. Data were collected and analysed using SPSS version 20. Results: A diploid chromosome number of 2n = 80 with an autosomal fundamental number (NFa) of 66 and 95 were obtained for the species of C. gambianus used in this study. The X chromosomes were medium-sized metacentric and small acrocentric while the Y chromosome was small acrocentric. Conclusion: Cricetomysgambianus was found to have an identifiable autosomal diploid number,The findings resembled those in Benin, Senegal, Niger, Cameroun and other countries. -

INSIGHTS INTO RELATIONSHIPS AMONG RODENT LINEAGES BASED on MITOCHONDRIAL GENOME SEQUENCE DATA a Dissertation by LAURENCE JOHN FR
INSIGHTS INTO RELATIONSHIPS AMONG RODENT LINEAGES BASED ON MITOCHONDRIAL GENOME SEQUENCE DATA A Dissertation by LAURENCE JOHN FRABOTTA Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY December 2005 Major Subject: Zoology INSIGHTS INTO RELATIONSHIPS AMONG RODENT LINEAGES BASED ON MITOCHONDRIAL GENOME SEQUENCE DATA A Dissertation by LAURENCE JOHN FRABOTTA Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Approved by: Chair of Committee, Rodney L. Honeycutt Committee Members, James B. Woolley John W. Bickham James R. Manhart Head of Department, Vincent M. Cassone December 2005 Major Subject: Zoology iii ABSTRACT Insights into Relationships among Rodent Lineages Based on Mitochondrial Genome Sequence Data. (December 2005) Laurence John Frabotta, B.S.; M.S., California State University, Long Beach Chair of Advisory Committee: Dr. Rodney L. Honeycutt This dissertation has two major sections. In Chapter II, complete mitochondrial (mt DNA) genome sequences were used to construct a hypothesis for affinities of most major lineages of rodents that arose quickly in the Eocene and were well established by the end of the Oligocene. Determining the relationships among extant members of such old lineages can be difficult. Two traditional schemes on subordinal classification of rodents have persisted for over a century, dividing rodents into either two or three suborders, with relationships among families or superfamilies remaining problematic. The mtDNA sequences for four new rodent taxa (Aplodontia, Cratogeomys, Erethizon, and Hystrix), along with previously published Euarchontoglires taxa, were analyzed under parsimony, likelihood, and Bayesian criteria. -

Micheal L. Dent Richard R. Fay Arthur N. Popper Editors Rodent Bioacoustics Springer Handbook of Auditory Research
Springer Handbook of Auditory Research Micheal L. Dent Richard R. Fay Arthur N. Popper Editors Rodent Bioacoustics Springer Handbook of Auditory Research Volume 67 Series Editor Richard R. Fay, Ph.D., Loyola University Chicago, Chicago, IL, USA Arthur N. Popper, Ph.D., University of Maryland, College Park, MD, USA Editorial Board Karen Avraham, Ph.D., Tel Aviv University, Israel Andrew Bass, Ph.D., Cornell University Lisa Cunningham, Ph.D., National Institutes of Health Bernd Fritzsch, Ph.D., University of Iowa Andrew Groves, Ph.D., Baylor University Ronna Hertzano, M.D., Ph.D., School of Medicine, University of Maryland Colleen Le Prell, Ph.D., University of Texas, Dallas Ruth Litovsky, Ph.D., University of Wisconsin Paul Manis, Ph.D., University of North Carolina Geoffrey Manley, Ph.D., University of Oldenburg, Germany Brian Moore, Ph.D., Cambridge University, UK Andrea Simmons, Ph.D., Brown University William Yost, Ph.D., Arizona State University More information about this series at http://www.springer.com/series/2506 The ASA Press The ASA Press imprint represents a collaboration between the Acoustical Society of America and Springer dedicated to encouraging the publication of important new books in acoustics. Published titles are intended to reflect the full range of research in acoustics. ASA Press books can include all types of books published by Springer and may appear in any appropriate Springer book series. Editorial Board Mark F. Hamilton (Chair), University of Texas at Austin James Cottingham, Coe College Diana Deutsch, University of California, San Diego Timothy F. Duda, Woods Hole Oceanographic Institution Robin Glosemeyer Petrone, Threshold Acoustics William M. -

Over 40% of All Mammal Species in the Next 2 Labs
Rodents Class Rodentia 5 (depends) Suborders 33 (maybe more) Families about 481 genera, 2277+ species Over 40% of all mammal species in the next 2 labs Sciuromorpha: squirrels, dormice, mountain beaver, and relatives Castorimorpha: beavers, gophers, kangaroo rats, pocket mice, and relatives Myomorpha: mice, rats, gerbils, jerboas, and relatives Anomaluromorpha: scaly-tailed squirrels and springhares Hystricomorpha: hystricognath rodents...lots of South American and African species, mostly Because rodents are such a Why rodents are evil... diverse and speciose group, their higher-level taxonomy keeps being revised. Hard to keep up! In recent decades, there have been 2, 3, 4 or 5 Suborders, depending on the revision, and Families keep getting pooled and split. We’ll just focus on some of the important Families and leave their relationships to future generations. They are a diverse and Why rodents are fun... speciose group, occur in just about every kind of habitat and climate, and show the broadest ecological diversity of any group of mammals. There are terrestrial, arboreal, scansorial, subterranean, and semiaquatic rodents. There are solitary, pair-forming, and social rodents. There are plantigrade, cursorial, You could spend your whole fossorial, bipedal, swimming life studying this group! and gliding rodents. (Some do.) General characteristics of rodents •Specialized ever-growing, self-sharpening incisors (2 upper, 2 lower) separated from cheek teeth by diastema; no canines •Cheek teeth may be ever-growing or rooted, but show a variety of cusp patterns, often with complex loops and folds of enamel and dentine reflecting the diet; cusp patterns also often useful taxonomically •Mostly small, average range of body size is 20-100 g, but some can get pretty large (capybara is largest extant species, may reach 50 kg) •Mostly herbivorous (including some specialized as folivores and granivores) or omnivorous •Females with duplex uterus, baculum present in males •Worldwide distribution, wide range of habitats and ecologies And now, on to a few Families.. -

Aplodontid, Sciurid, Castorid, Zapodid and Geomyoid Rodents of the Rodent Hill Locality, Cypress Hills Formation, Southwest Saskatchewan
APLODONTID, SCIURID, CASTORID, ZAPODID AND GEOMYOID RODENTS OF THE RODENT HILL LOCALITY, CYPRESS HILLS FORMATION, SOUTHWEST SASKATCHEWAN A Thesis Submitted to the College of Graduate Studies and Research in Partial Fulfillment of the Requirements for the Degree of Master of Science in the Department of Geological Sciences University of Saskatchewan Saskatoon By Sean D. Bell © Copyright Sean D. Bell, December 2004. All rights reserved. PERMISSION TO USE In presenting this thesis in partial fulfilment of the requirements for a Master’s degree from the University of Saskatchewan, I agree that the libraries of the University of Saskatchewan may make it freely available for inspection. I further agree that permission for copying of this thesis in any manner, in whole or in part, for scholarly purposes may be granted by the professors who supervised my thesis work or, in their absence, by the Head of the Department of Geological Sciences or the Dean of the College of Graduate Studies and Research. It is understood that any copying or publication or use of this thesis or parts thereof for financial gain shall not be allowed without my written permission. It is also understood that due recognition shall be given to me and to the University of Saskatchewan in any scholarly use which may be made of any material in my thesis. Requests for permission to copy or to make other use of material in this thesis in whole or part should be addressed to: Head of the Department of Geological Sciences 114 Science Place University of Saskatchewan Saskatoon, Saskatchewan S7N 5E2 i ABSTRACT The Rodent Hill Locality is a fossil-bearing site that is part of the Cypress Hills Formation, and is located roughly 15 km northwest of the town of Eastend, Saskatchewan. -

Phylogenetic Relationships in Neotomine-Peromyscine Rodents (Muroidea) and a Reappraisal of the Dichotomy Within New World Cricetinae
MISCELLANEOUS PUBLICATIONS MUSEUM OF ZOOLOGY, UNIVERSITY OF MICHIGAN NO. 157 Phylogenetic Relationships in Neotomine-Peromyscine Rodents (Muroidea) and a Reappraisal of the Dichotomy within New World Cricetinae by Michael Dean Carleton National Museum of Natural History Smithsonian Institution Washington, D.C. 20560 Ann Arbor MUSEUM OF ZOOLOGY, UNIVERSITY OF MICHIGAN December 12, 1980 MISCELLANEOUS PUBLICATIONS MUSEUM OF ZOOLOGY, UNIVERSITY OF MICHIGAN The publications of the Museum of Zoology, University of Michigan, consist of two series - the Occasional Papers and the Miscellaneous Publications. Both series were founded by Dr. Bryant Walker, Mr. Bradshaw H. Swales, and Dr. W. W. Newcomb. The Occasional Papers, publication of which was begun in 1913, serve as a medium for original studies based principally upon the collections in the Museum. They are issued separately. When a sufficient number bf pages has been printed to make a volume, a title page, table of contents, and an index are supplied to libraries and individuals on the mail- ing list for the series. The Miscellaneous Publications. which include .DaDers . on field and museum tech- niques, monographic studies, and other contributions not within the scope of the Occa- sional Papers, are published separately. It is not intended that they be grouped into volumes. Each number has a title page and, when necessary, a table of contents. A complete list of publications on Birds, Fishes, Insects, Mammals, Mollusks, and Reptiles and Amphibians is available. Address inquiries to the Director, Museum of Zool- ogy, Ann Arbor, Michigan 48109. MISCELLANEOUS PUBLICATIONS MUSEUM OF ZOOLOGY, UNIVERSITY OF MICHIGAN NO. 157 Phylogenetic Relationships in Neotomine-Peromyscine Rodents (Muroidea) and a Reappraisal of the Dichotomy within New World Cricetinae hy Michael Dean Carleton National Museum of Natural History Smithsonian Institution Washington, D.C. -

Phylogenetic Relationships and Divergence Times in Rodents Based on Both Genes and Fossils Ryan Norris University of Vermont
University of Vermont ScholarWorks @ UVM Graduate College Dissertations and Theses Dissertations and Theses 2009 Phylogenetic Relationships and Divergence Times in Rodents Based on Both Genes and Fossils Ryan Norris University of Vermont Follow this and additional works at: https://scholarworks.uvm.edu/graddis Recommended Citation Norris, Ryan, "Phylogenetic Relationships and Divergence Times in Rodents Based on Both Genes and Fossils" (2009). Graduate College Dissertations and Theses. 164. https://scholarworks.uvm.edu/graddis/164 This Dissertation is brought to you for free and open access by the Dissertations and Theses at ScholarWorks @ UVM. It has been accepted for inclusion in Graduate College Dissertations and Theses by an authorized administrator of ScholarWorks @ UVM. For more information, please contact [email protected]. PHYLOGENETIC RELATIONSHIPS AND DIVERGENCE TIMES IN RODENTS BASED ON BOTH GENES AND FOSSILS A Dissertation Presented by Ryan W. Norris to The Faculty of the Graduate College of The University of Vermont In Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Specializing in Biology February, 2009 Accepted by the Faculty of the Graduate College, The University of Vermont, in partial fulfillment of the requirements for the degree of Doctor of Philosophy, specializing in Biology. Dissertation ~xaminationCommittee: w %amB( Advisor 6.William ~il~atrickph.~. Duane A. Schlitter, Ph.D. Chairperson Vice President for Research and Dean of Graduate Studies Date: October 24, 2008 Abstract Molecular and paleontological approaches have produced extremely different estimates for divergence times among orders of placental mammals and within rodents with molecular studies suggesting a much older date than fossils. We evaluated the conflict between the fossil record and molecular data and find a significant correlation between dates estimated by fossils and relative branch lengths, suggesting that molecular data agree with the fossil record regarding divergence times in rodents. -

Rodentia (Sciuromorpha and Castorimorpha) in the National Museum of Natural History, Smithsonian Institution
Smithsonian Institution Scholarly Press SMITHSONIAN CONTRIBUTIONS TO ZOOLOGY • NUMBER 640 Catalog of Type Specimens of Recent Mammals: Rodentia (Sciuromorpha and Castorimorpha) in the National Museum of Natural History, Smithsonian Institution Robert D. Fisher and Craig A. Ludwig Contents INTRODUCTION 1 Type Categories Included 1 Organizatioii 2 Formant of Accounts 2 Approach 2 Acknowledgments 3 ÁCCOUNTS OF NAME-BEARING TYPE SPECIMENS 3 Suborder Sciuromorpha Brandt, 1855 3 Family Aplodontiidae Brandt, 1855 3 Genus Aplodontia Richardsoii, 1829 3 Family Sciuridae G. Fischer, 1814 3 Genus Aretes G. M. Alien, 1940 3 Genus Ammospermophilus Merriam, 1892 4 Genus Callosdurus Gray, 1867 4 Genus Callospermophilus, Merriam, 1897 10 Gemís Cynomys Rafinesque, 1817 11 Genus Dremomys Heude, 1898 11 Genus Exilisdurifs Moore, 1958 11 Genus Glaucomys Thomas, 1908 11 Genus Heliosdurus Trouessart, 1880 13 Genus Hylopetes Thomas, 1908 13 Genus Ictidomys ]. A. Alien, 1877 13 Genus lomys Thomas, 1908 14 Genus Lariscus Thomas and Wroughton, 1909 14 Genus Marmota Blumenbach, 1779 14 Gemís Menetes Thomas, 1908 15 Genus Microsdurus J. A. Alien, 'i 895 15 Genus Nannosdurus Trouessart, 1880 16 Genus Notodtellus A. H. Howell, 1938 16 Genus Otospermophilus Brandt, 1844 16 Genus Paraxerus Forsyth Major, 1893 17 Genus Petaurista Link, 1795 17 SMITHSONIAN CONTRIBUTIONS TO ZOOLOGY Genus Petinomys Thomas, 1908 17 Genus Prosciurillus Ellerman, 1947 17 Genus Protoxerus Forsyth Major, 1893 18 Genus Pteromys G. Cuvier, 1800 18 Genus Ratufa Gray, 1867 18 Genus Sdurus Linnaeus, -

New Records of Rodentia from the Duchesnean (Middle Eocene) Simi Valley Landfill Local Fauna, Sespe Formation, California
Paludicola 8(1):49-73 September 2010 © by the Rochester Institute of Vertebrate Paleontology NEW RECORDS OF RODENTIA FROM THE DUCHESNEAN (MIDDLE EOCENE) SIMI VALLEY LANDFILL LOCAL FAUNA, SESPE FORMATION, CALIFORNIA Thomas S. Kelly Research Associate, Vertebrate Paleontology Section, Natural History Museum of Los Angeles County, 900 Exposition Blvd, Los Angeles, California 90007 ABSTRACT A large number of isolated rodent teeth have been recently recovered from the Duchesnean (middle Eocene) Simi Valley Landfill Local Fauna of the Sespe Formation during a paleontologic mitigation program at the Simi Valley Landfill and Recycling Center, Ventura County, California. Included in these teeth are new samples of Metanoiamys korthi, Paradjidaumo reynoldsi, Simiacritomys whistleri, and Simimys landeri. The discovery of upper and putative lower premolars of Simiacritomys whistleri supports it’s referral to Eomyidae. New occurrences for the Simi Valley Landfill Local Fauna include Eomyidae (one or more species of uncertain affinities) and Pareumys sp. INTRODUCTION Within the new specimens from bed 30A are numerous additional rodent teeth, including those of The middle member of the Sespe Formation, species that were previously known from small sample which is exposed along the north side of Simi Valley, sizes (Metanoiamys korthi Kelly and Whistler, 1998, Ventura County, California, has previously yielded Paradjidaumo reynoldsi Kelly, 1992, Simiacritomys numerous middle Eocene (Uintan and Duchesnean) whistleri Kelly, 1992, and Simimys landeri Kelly, fossil mammals (e.g., Golz, 1976; Golz and 1992). Also, included within these specimens are the Lillegraven, 1977; Mason, 1988; Kelly, 1990, 1992, first records of Pareumys sp. and at least one or more 2009, 2010; Kelly et al., 1991; Kelly and Whistler, undetermined eomyid species from bed 30A. -
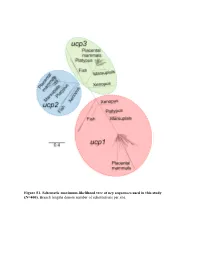
Figure S1. Schematic Maximum-Likelihood Tree of Ucp Sequences Used in This Study (N=400)
Figure S1. Schematic maximum-likelihood tree of ucp sequences used in this study (N=400). Branch lengths denote number of substitutions per site. Exon 1 1 10 20 30 40 50 |...|....|....|....|....|....|....|....|....|....| Homo sapiens ATGGGGGGCCTGA-CAGCCTCGGACGTACACCCGACCC---TGGGGGTCC Bradypus variegatus ?????????????????????????????????????????????????? Choloepus hoffmanni ?????????????????????????????????????????????????? Mylodon darwinii ?????????????????????????????????????????????????? Cyclopes didactylus ?????????????????????????????????????????????????? Dasypus novemcinctus ATGGGGCGCCAGGGCTCCCGCGGGCTCACCCCCCGC-------------- Procavia capensis CTG--AGTTAAGA-CAACCTCAGAAATGCCGCCTAC-----------GCA Elephas maximus ACGGTAGGCCAGA-CGACCGCAGACGTGCCCCGGACCATGGTGGGGGTCA Mammuthus primigenius ACGGTAGGCCAGA-CGACCGCAGACGTGCCCCGGACCATGGTGGGGGTCA Loxodonta africana ACCGTAGGCCAGA-CGACCGCAGACGTGCCCCGGACCATGGTGGGGGTCA Trichechus manatus ATGGTGGGCCAGA-CTACCTGGGATGTGCCCCCGACCA---TGGGCGTCA Dugong dugon ATGGTGGGCCAGA-CTACCTCGGATGTGCCCCCGACCA---TGGGCGTCA Hydrodamalis gigas ATGGTGGGCCAGA-CTACCTCGGATGTGCCCCCGACCA---TGGGCGTCA Sus scrofa CTGTCAGGA-TGA-CAGTTCCTGAAGTGCCCCCGACCA---TAGCGGTCA Sus verrucosus CTGTCAGGA-TGA-CAGTTCCTGAAGTGCCCTCGACCA---TAGCGGTCA Sus cebifrons CTGTCAGGA-TGA-CAGTTCCTGAAGTGCCCTCGACCA---TAGCGGTCA Physeter macrocephalus ATGGTGGGACTCG-CAGCCTCATACGTGCCCCCGACCA---TGGCGGTCA Delphinapterus leucas -------------------------------------------------- Lipotes vexillifer ATGGTGGGACTCG-CAGCCTCAGACGTGCCCCCGACCA---TGGCGGTCA Balaena mysticetus ATGGTGGCACTCA-CAGCCTCAGACGTGCCCCCGACCA---TGGCGGTCA