Micheal L. Dent Richard R. Fay Arthur N. Popper Editors Rodent Bioacoustics Springer Handbook of Auditory Research
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Auditory-Vocal Coupling in the Naked Mole-Rat, a Mammal with Poor Auditory Thresholds
Journal of Comparative Physiology A https://doi.org/10.1007/s00359-018-1287-8 ORIGINAL PAPER Auditory-vocal coupling in the naked mole-rat, a mammal with poor auditory thresholds Kazuo Okanoya1,2 · Shigeto Yosida2 · Catherine M. Barone3 · Daniel T. Applegate3 · Elizabeth F. Brittan‑Powell4 · Robert J. Dooling4 · Thomas J. Park3 Received: 7 May 2018 / Revised: 4 September 2018 / Accepted: 7 September 2018 © The Author(s) 2018 Abstract Naked mole-rats are extremely social and extremely vocal rodents, displaying a wide range of functionally distinct call types and vocalizing almost continuously. Their vocalizations are low frequency, and a behavioral audiogram has shown that naked mole-rats, like other subterranean mammals, hear only low frequencies. Hence, the frequency range of their hearing and vocalizations appears to be well matched. However, even at low frequencies, naked mole-rats show very poor auditory thresholds, suggesting vocal communication may be effective only over short distances. However, in a tunnel environment where low frequency sounds propagate well and background noise is low, it may be that vocalizations travel considerable distances at suprathreshold intensities. Here, we confirmed hearing sensitivity using the auditory brainstem response; we characterized signature and alarm calls in intensity and frequency domains and we measured the effects of propagation through tubes with the diameter of naked mole-rat tunnels. Signature calls—used for intimate communication—could travel 3–8 m at suprathreshold intensities, and alarm calls (lower frequency and higher intensity), could travel up to 15 m. Despite this species’ poor hearing sensitivity, the naked mole-rat displays a functional, coupled auditory-vocal communication system—a hallmark principle of acoustic communication systems across taxa. -

An Ecological Comparison of Small Mammal Communities in California
An Ecological Comparison of Small Mammal Abstract: Our studies in similar scrub habitats Communities in California and Chile1 in California and Chile reveal some interesting differences between these two regions in the structure of their small mammal communities. The Chilean fauna is less diverse, with fewer species 2 William E. Glanz and Peter L. Meserve per site, and possibly more extreme density fluc- tuations. Chilean rodents are more strongly associated with areas of high shrub and rock cover, while California species show a greater variety of habitat preferences. Chile has more insectivorous species, and California has more seed-eating specialists. Some of these differ- ences may be related to biogeographic and climatic factors, while others may reflect a longer history of human disturbance in Chile. Similar climates often seem to favor the evo- FIELD SITES AND METHODS lution of similar organisms, and the Mediterranean -climate regions of the world have provided numer- We have ecological data from a variety of ous examples of such evolutionary convergence. communities in each region, but will restrict our Of the five regions with this climate, California discussion here to results from three vegetational and Chile have been compared very extensively types which we have studied intensively: dry (Mooney 1977), particularly in terms of the mor- coastal scrub, moist coastal scrub, and evergreen phological, physiological, and ecological attri- chaparral. For each of these, the vegetation butes of their dominant organisms. Striking structure and life forms at our sites were closely similarities between these two regions have been matched between continents. found in their vegetation structure and community patterns (Mooney and Dunn 1970; Parsons and The dry coastal scrub localities were studied Moldenke 1975; Parsons 1976), their birds (Cody by Meserve near Irvine, Orange County, California 1973 and 1974), and their lizards (Fuentes 1976). -

Geographic Distribution of Hantaviruses Associated with Neotomine and Sigmodontine Rodents, Mexico Mary L
Geographic Distribution of Hantaviruses Associated with Neotomine and Sigmodontine Rodents, Mexico Mary L. Milazzo,1 Maria N.B. Cajimat,1 Hannah E. Romo, Jose G. Estrada-Franco, L. Ignacio Iñiguez-Dávalos, Robert D. Bradley, and Charles F. Fulhorst To increase our knowledge of the geographic on the North American continent are Bayou virus, Black distribution of hantaviruses associated with neotomine or Creek Canal virus (BCCV), Choclo virus (CHOV), New sigmodontine rodents in Mexico, we tested 876 cricetid York virus, and Sin Nombre virus (SNV) (3–7). Other rodents captured in 18 Mexican states (representing at hantaviruses that are principally associated with neotomine least 44 species in the subfamily Neotominae and 10 or North American sigmodontine rodents include Carrizal species in the subfamily Sigmodontinae) for anti-hantavirus virus (CARV), Catacamas virus, El Moro Canyon virus IgG. We found antibodies against hantavirus in 35 (4.0%) rodents. Nucleotide sequence data from 5 antibody-positive (ELMCV), Huitzilac virus (HUIV), Limestone Canyon rodents indicated that Sin Nombre virus (the major cause of virus (LSCV), Montano virus (MTNV), Muleshoe virus hantavirus pulmonary syndrome [HPS] in the United States) (MULV), Playa de Oro virus, and Rio Segundo virus is enzootic in the Mexican states of Nuevo León, San Luis (RIOSV) (8–14). Potosí, Tamaulipas, and Veracruz. However, HPS has not Specifi c rodents (usually 1 or 2 closely related been reported from these states, which suggests that in species) are the principal hosts of the hantaviruses, northeastern Mexico, HPS has been confused with other for which natural host relationships have been well rapidly progressive, life-threatening respiratory diseases. -

Special Publications Museum of Texas Tech University Number 63 18 September 2014
Special Publications Museum of Texas Tech University Number 63 18 September 2014 List of Recent Land Mammals of Mexico, 2014 José Ramírez-Pulido, Noé González-Ruiz, Alfred L. Gardner, and Joaquín Arroyo-Cabrales.0 Front cover: Image of the cover of Nova Plantarvm, Animalivm et Mineralivm Mexicanorvm Historia, by Francisci Hernández et al. (1651), which included the first list of the mammals found in Mexico. Cover image courtesy of the John Carter Brown Library at Brown University. SPECIAL PUBLICATIONS Museum of Texas Tech University Number 63 List of Recent Land Mammals of Mexico, 2014 JOSÉ RAMÍREZ-PULIDO, NOÉ GONZÁLEZ-RUIZ, ALFRED L. GARDNER, AND JOAQUÍN ARROYO-CABRALES Layout and Design: Lisa Bradley Cover Design: Image courtesy of the John Carter Brown Library at Brown University Production Editor: Lisa Bradley Copyright 2014, Museum of Texas Tech University This publication is available free of charge in PDF format from the website of the Natural Sciences Research Laboratory, Museum of Texas Tech University (nsrl.ttu.edu). The authors and the Museum of Texas Tech University hereby grant permission to interested parties to download or print this publication for personal or educational (not for profit) use. Re-publication of any part of this paper in other works is not permitted without prior written permission of the Museum of Texas Tech University. This book was set in Times New Roman and printed on acid-free paper that meets the guidelines for per- manence and durability of the Committee on Production Guidelines for Book Longevity of the Council on Library Resources. Printed: 18 September 2014 Library of Congress Cataloging-in-Publication Data Special Publications of the Museum of Texas Tech University, Number 63 Series Editor: Robert J. -

Wild Patagonia & Central Chile
WILD PATAGONIA & CENTRAL CHILE: PUMAS, PENGUINS, CONDORS & MORE! NOVEMBER 1–18, 2019 Pumas simply rock! This year we enjoyed 9 different cats! Observing the antics of lovely Amber here and her impressive family of four cubs was certainly the highlight in Torres del Paine National Park — Photo: Andrew Whittaker LEADERS: ANDREW WHITTAKER & FERNANDO DIAZ LIST COMPILED BY: ANDREW WHITTAKER VICTOR EMANUEL NATURE TOURS, INC. 2525 WALLINGWOOD DRIVE, SUITE 1003 AUSTIN, TEXAS 78746 WWW.VENTBIRD.COM Sensational, phenomenal, outstanding Chile—no superlatives can ever adequately describe the amazing wildlife spectacles we enjoyed on this year’s tour to this breathtaking and friendly country! Stupendous world-class scenery abounded with a non-stop array of exciting and easy birding, fantastic endemics, and super mega Patagonian specialties. Also, as I promised from day one, everyone fell in love with Chile’s incredible array of large and colorful tapaculos; we enjoyed stellar views of all of the country’s 8 known species. Always enigmatic and confiding, the cute Chucao Tapaculo is in the Top 5 — Photo: Andrew Whittaker However, the icing on the cake of our tour was not birds but our simply amazing Puma encounters. Yet again we had another series of truly fabulous moments, even beating our previous record of 8 Pumas on the last day when I encountered a further 2 young Pumas on our way out of the park, making it an incredible 9 different Pumas! Our Puma sightings take some beating, as they have stood for the last three years at 6, 7, and 8. For sure none of us will ever forget the magical 45 minutes spent observing Amber meeting up with her four 1- year-old cubs as they joyfully greeted her return. -

Comparative Analyses of Past Population Dynamics Between Two Subterranean Zokor Species and the Response to Climate Changes
Turkish Journal of Zoology Turk J Zool (2013) 37: 143-148 http://journals.tubitak.gov.tr/zoology/ © TÜBİTAK Research Article doi:10.3906/zoo-1204-18 Comparative analyses of past population dynamics between two subterranean zokor species and the response to climate changes 1 1 1 1 1 1 Lizhou TANG , Long YU , Weidong LU , Junjie WANG , Mei MA , Xiaodong SHI , 1, 2 Jiangang CHEN *, Tongzuo ZHANG 1 College of Biologic Resource and Environmental Science, Qujing Normal University, Qujing, Yunnan 655011, China 2 Key Laboratory of the Qinghai-Tibetan Plateau Ecosystem and Biological Evolution and Adaptation, Northwest Institute of Plateau Biology, Chinese Academy of Sciences, Xining, Qinghai 810001, China Received: 16.04.2012 Accepted: 21.09.2012 Published Online: 25.02.2013 Printed: 25.03.2013 Abstract: The mitochondrial cytochrome b sequences of 34 haplotypes from 114 individuals of Eospalax baileyi and GenBank data of 40 haplotypes from 121 individuals of Eospalax cansus were used to investigate if the Quaternary glaciations on the Qinghai-Tibetan Plateau (QTP) influenced the distribution and colonisation of these 2 species, and if both species presented congruous past population dynamics. The phylogenetic tree indicated a valid status for the genera Eospalax and Myospalax, and an independent species status for E. baileyi and E. cansus. The demographical population history of E. baileyi showed that the effective population size remained stable between 1.00 and 0.50 million years ago (Mya), increased quickly to a peak and fluctuated dramatically between 0.50 and 0.20 Mya, after which a stable level was maintained from 0.20 Mya to the present. -

Transcriptome Analysis of the Liver of Eospalax Fontanierii Under Hypoxia
Transcriptome Analysis of the Liver of Eospalax Fontanierii Under Hypoxia Zhiqiang Hao Shaanxi Normal University College of Life Sciences Lulu Xu Shaanxi Normal University College of Life Sciences Jianping He Shaanxi Normal University College of Life Sciences Guanglin Li Shaanxi Normal University College of Life Sciences Jingang Li ( [email protected] ) Shaanxi Normal University https://orcid.org/0000-0002-5664-0715 Research Keywords: Hypoxia adaptation, Eospalax fontanierii, transcriptome Posted Date: September 15th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-74186/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/27 Abstract Background Hypoxia can induce cell damage, inammation, carcinogenesis, and inhibit liver regeneration in non- adapted species. Because of their excellent hypoxia adaptation features, subterranean rodents have been widely studied to clarify the mechanism of hypoxia adaptation. Eospalax fontanierii, which is a subterranean rodent found in China, can survive for more than 10 h under 4% O2 without observable injury, while Sprague-Dawle rats can survive for less than 6 h under the same conditions. To explore the potential mechanism of hypoxia adaptation in E. fontanierii, we performed RNA-seq analysis of the liver in E. fontanierii exposed to different oxygen levels (6.5%, 10.5%, and 21%). Results Based on the bioinformatics analysis, 39,439 unigenes were assembled, and 56.78% unigenes were annotated using public databases (Nr, GO, Swiss-Prot, KEGG, and Pfam). In total, 725 differentially expressed genes (DEGs) were identied in the response to hypoxia; six with important functions were validated by qPCR. -

Downloaded from Ensembl (Www
Lin et al. BMC Genomics 2014, 15:32 http://www.biomedcentral.com/1471-2164/15/32 RESEARCH ARTICLE Open Access Transcriptome sequencing and phylogenomic resolution within Spalacidae (Rodentia) Gong-Hua Lin1, Kun Wang2, Xiao-Gong Deng1,3, Eviatar Nevo4, Fang Zhao1, Jian-Ping Su1, Song-Chang Guo1, Tong-Zuo Zhang1* and Huabin Zhao5* Abstract Background: Subterranean mammals have been of great interest for evolutionary biologists because of their highly specialized traits for the life underground. Owing to the convergence of morphological traits and the incongruence of molecular evidence, the phylogenetic relationships among three subfamilies Myospalacinae (zokors), Spalacinae (blind mole rats) and Rhizomyinae (bamboo rats) within the family Spalacidae remain unresolved. Here, we performed de novo transcriptome sequencing of four RNA-seq libraries prepared from brain and liver tissues of a plateau zokor (Eospalax baileyi) and a hoary bamboo rat (Rhizomys pruinosus), and analyzed the transcriptome sequences alongside a published transcriptome of the Middle East blind mole rat (Spalax galili). We characterize the transcriptome assemblies of the two spalacids, and recover the phylogeny of the three subfamilies using a phylogenomic approach. Results: Approximately 50.3 million clean reads from the zokor and 140.8 million clean reads from the bamboo ratwere generated by Illumina paired-end RNA-seq technology. All clean reads were assembled into 138,872 (the zokor) and 157,167 (the bamboo rat) unigenes, which were annotated by the public databases: the Swiss-prot, Trembl, NCBI non-redundant protein (NR), NCBI nucleotide sequence (NT), Gene Ontology (GO), Cluster of Orthologous Groups (COG), and Kyoto Encyclopedia of Genes and Genomes (KEGG). -

Inventory of Mammals at Walnut Canyon, Wupatki, and Sunset Crater National Monuments
National Park Service U.S. Department of the Interior Natural Resource Program Center Inventory of Mammals at Walnut Canyon, Wupatki, and Sunset Crater National Monuments Natural Resource Technical Report NPS/SCPN/NRTR–2009/278 ON THE COVER: Top: Wupatki National Monument; bottom left: bobcat (Lynx rufus); bottom right: Wupatki pocket mouse (Perogna- thus amplus cineris) at Wupatki National Monument. Photos courtesy of U.S. Geological Survey/Charles Drost. Inventory of Mammals at Walnut Canyon, Wupatki, and Sunset Crater National Monuments Natural Resource Technical Report NPS/SCPN/NRTR—2009/278 Author Charles Drost U.S. Geological Survey Southwest Biological Science Center 2255 N. Gemini Drive Flagstaff, AZ 86001 Editing and Design Jean Palumbo National Park Service, Southern Colorado Plateau Network Northern Arizona University Flagstaff, Arizona December 2009 U.S. Department of the Interior National Park Service Natural Resource Program Center Fort Collins, Colorado The National Park Service, Natural Resource Program Center publishes a range of reports that address natural resource topics of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Technical Report Series is used to disseminate results of scientific studies in the physical, biological, and social sciences for both the advancement of science and the achievement of the National Park Service mission. The series provides contributors with a forum for displaying comprehensive data that are often deleted from journals because of page limitations. All manuscripts in the series receive the appropriate level of peer review to ensure that the information is scientifically credible, technically accurate, appropriately written for the intended audience, and designed and published in a professional manner. -
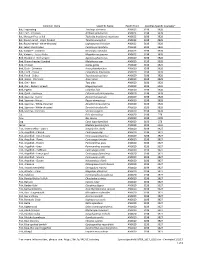
*For More Information, Please See
Common Name Scientific Name Health Point Specifies-Specific Course(s)* Bat, Frog-eating Trachops cirrhosus AN0023 3198 3928 Bat, Fruit - Jamaican Artibeus jamaicensis AN0023 3198 3928 Bat, Mexican Free-tailed Tadarida brasiliensis mexicana AN0023 3198 3928 Bat, Round-eared - stripe-headed Tonatia saurophila AN0023 3198 3928 Bat, Round-eared - white-throated Lophostoma silvicolum AN0023 3198 3928 Bat, Seba's short-tailed Carollia perspicillata AN0023 3198 3928 Bat, Vampire - Common Desmodus rotundus AN0023 3198 3928 Bat, Vampire - Lesser False Megaderma spasma AN0023 3198 3928 Bird, Blackbird - Red-winged Agelaius phoeniceus AN0020 3198 3928 Bird, Brown-headed Cowbird Molothurus ater AN0020 3198 3928 Bird, Chicken Gallus gallus AN0020 3198 3529 Bird, Duck - Domestic Anas platyrhynchos AN0020 3198 3928 Bird, Finch - House Carpodacus mexicanus AN0020 3198 3928 Bird, Finch - Zebra Taeniopygia guttata AN0020 3198 3928 Bird, Goose - Domestic Anser anser AN0020 3198 3928 Bird, Owl - Barn Tyto alba AN0020 3198 3928 Bird, Owl - Eastern Screech Megascops asio AN0020 3198 3928 Bird, Pigeon Columba livia AN0020 3198 3928 Bird, Quail - Japanese Coturnix coturnix japonica AN0020 3198 3928 Bird, Sparrow - Harris' Zonotrichia querula AN0020 3198 3928 Bird, Sparrow - House Passer domesticus AN0020 3198 3928 Bird, Sparrow - White-crowned Zonotrichia leucophrys AN0020 3198 3928 Bird, Sparrow - White-throated Zonotrichia albicollis AN0020 3198 3928 Bird, Starling - Common Sturnus vulgaris AN0020 3198 3928 Cat Felis domesticus AN0020 3198 279 Cow Bos taurus -

Blueprint Earth Field Guide
Blueprint Earth Field Guide Plants Note that this list is not comprehensive. If you are uncertain of the identification you’ve made of a particular plant, take a picture and a voucher (when possible) and discuss your observations with the Supervisory Scientist team. Trees & Bushes Joshua tree - Yucca brevifolia Parry saltbush - Atriplex parryi Mojave sage - Salvia pachyphylla Creosote bush - Larrea tridentata Mojave yucca - Yucca schidigera Chaparral yucca - Yucca whipplei Torr. Desert holly - A. hymenelytra Torr. Manzanita - Arctostaphylos Adans. Cacti Barrel cactus - Ferocactus cylindraceus var. Jumping cholla - Cylindropuntia bigelovii Engelm. lecontei Foxtail cactus - Escobaria vivipara var. alversonii Silver cholla - Opuntia echinocarpa var. echinocarpa Pencil cholla - Opuntia ramosissima Cottontop cactus - Echinocactus polycephalus Hedgehog cactus - Echinocereus engelmanii var. Mojave mound cactus - Echinocereeus chrysocentrus triglochiderus var. mojavensis Beavertail cactus - Opuntia basilaris Grasses Indian Rice Grass - Oryzopsis hymenoides Bush Muhly - Muhlenbergia porteri Fluff Grass - Erioneuron pulchella Red Brome - Bromus rubens Desert Needle - Stipa speciosa Big Galleta – Hilaria rigida Flowers Wooly Amsonia Chuparosa Amsonia tomentosa Justicia californica Brittlebush Encelia farinosa Chia Salvia columbariae Sacred Datura Desert Calico Datura wrightii Loeseliastrum matthewsii Bigelow Coreopsis Desert five-spot Coreopsis bigelovii Eremalche rotundifolia - Desert Chicory Rafinesquia Desert Lupine neomexicana Desert Larkspur -

Tissue Specific Regulatory Network Annotation for Non-Coding Elements
Quantitative Biology 2020, 8(1): 43–50 https://doi.org/10.1007/s40484-020-0195-4 RESEARCH ARTICLE ZokorDB: tissue specificregulatorynetwork annotation for non-coding elements of plateau zokor Jingxue Xin1,6,7,†, Junjun Hao2,†, Lang Chen1, Tao Zhang3, Lei Li1,5,7, Luonan Chen3,5, Wenmin Zhao4, Xuemei Lu2,5, Peng Shi2,5,*, Yong Wang1,5,7,* 1 CEMS, NCMIS, MDIS, Academy of Mathematics and Systems Science, Chinese Academy of Sciences, Beijing 100190, China 2 State Key Laboratory of Genetic Resources and Evolution, Laboratory of Evolutionary and Functional Genomics, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650223, China 3 Key Laboratory of Systems Biology, Innovation Center for Cell Signaling Network, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200031, China 4 Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China 5 Center for Excellence in Animal Evolution and Genetics, Chinese Academy of Sciences, Kunming 650223, China 6 Department of Statistics, Department of Biomedical Data Science, Bio-X Program, Stanford University, Stanford, CA 94305, USA 7 University of Chinese Academy of Sciences, Beijing 100049, China * Correspondence: [email protected], [email protected] Received September 4, 2019; Revised December 16, 2019; Accepted December 23, 2019 Background: Plateau zokor inhabits in sealed burrows from 2,000 to 4,200 meters at Qinghai-Tibet Plateau. This extreme living environment makes it a great model to study animal adaptation to hypoxia, low temperature, and high carbon dioxide concentration. Methods: We provide an integrated resource, ZokorDB, for tissue specific regulatory network annotation for zokor.