Suffix Trees and Suffix Arrays in Primary and Secondary Storage Pang Ko Iowa State University
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

FM-Index Reveals the Reverse Suffix Array
FM-Index Reveals the Reverse Suffix Array Arnab Ganguly Department of Computer Science, University of Wisconsin - Whitewater, WI, USA [email protected] Daniel Gibney Department of Computer Science, University of Central Florida, Orlando, FL, USA [email protected] Sahar Hooshmand Department of Computer Science, University of Central Florida, Orlando, FL, USA [email protected] M. Oğuzhan Külekci Informatics Institute, Istanbul Technical University, Turkey [email protected] Sharma V. Thankachan Department of Computer Science, University of Central Florida, Orlando, FL, USA [email protected] Abstract Given a text T [1, n] over an alphabet Σ of size σ, the suffix array of T stores the lexicographic order of the suffixes of T . The suffix array needs Θ(n log n) bits of space compared to the n log σ bits needed to store T itself. A major breakthrough [FM–Index, FOCS’00] in the last two decades has been encoding the suffix array in near-optimal number of bits (≈ log σ bits per character). One can decode a suffix array value using the FM-Index in logO(1) n time. We study an extension of the problem in which we have to also decode the suffix array values of the reverse text. This problem has numerous applications such as in approximate pattern matching [Lam et al., BIBM’ 09]. Known approaches maintain the FM–Index of both the forward and the reverse text which drives up the space occupancy to 2n log σ bits (plus lower order terms). This brings in the natural question of whether we can decode the suffix array values of both the forward and the reverse text, but by using n log σ bits (plus lower order terms). -

Compressed Suffix Trees with Full Functionality
Compressed Suffix Trees with Full Functionality Kunihiko Sadakane Department of Computer Science and Communication Engineering, Kyushu University Hakozaki 6-10-1, Higashi-ku, Fukuoka 812-8581, Japan [email protected] Abstract We introduce new data structures for compressed suffix trees whose size are linear in the text size. The size is measured in bits; thus they occupy only O(n log |A|) bits for a text of length n on an alphabet A. This is a remarkable improvement on current suffix trees which require O(n log n) bits. Though some components of suffix trees have been compressed, there is no linear-size data structure for suffix trees with full functionality such as computing suffix links, string-depths and lowest common ancestors. The data structure proposed in this paper is the first one that has linear size and supports all operations efficiently. Any algorithm running on a suffix tree can also be executed on our compressed suffix trees with a slight slowdown of a factor of polylog(n). 1 Introduction Suffix trees are basic data structures for string algorithms [13]. A pattern can be found in time proportional to the pattern length from a text by constructing the suffix tree of the text in advance. The suffix tree can also be used for more complicated problems, for example finding the longest repeated substring in linear time. Many efficient string algorithms are based on the use of suffix trees because this does not increase the asymptotic time complexity. A suffix tree of a string can be constructed in linear time in the string length [28, 21, 27, 5]. -

1 Suffix Trees
This material takes about 1.5 hours. 1 Suffix Trees Gusfield: Algorithms on Strings, Trees, and Sequences. Weiner 73 “Linear Pattern-matching algorithms” IEEE conference on automata and switching theory McCreight 76 “A space-economical suffix tree construction algorithm” JACM 23(2) 1976 Chen and Seifras 85 “Efficient and Elegegant Suffix tree construction” in Apos- tolico/Galil Combninatorial Algorithms on Words Another “search” structure, dedicated to strings. Basic problem: match a “pattern” (of length m) to “text” (of length n) • goal: decide if a given string (“pattern”) is a substring of the text • possibly created by concatenating short ones, eg newspaper • application in IR, also computational bio (DNA seqs) • if pattern avilable first, can build DFA, run in time linear in text • if text available first, can build suffix tree, run in time linear in pattern. • applications in computational bio. First idea: binary tree on strings. Inefficient because run over pattern many times. • fractional cascading? • realize only need one character at each node! Tries: • used to store dictionary of strings • trees with children indexed by “alphabet” • time to search equal length of query string • insertion ditto. • optimal, since even hashing requires this time to hash. • but better, because no “hash function” computed. • space an issue: – using array increases stroage cost by |Σ| – using binary tree on alphabet increases search time by log |Σ| 1 – ok for “const alphabet” – if really fussy, could use hash-table at each node. • size in worst case: sum of word lengths (so pretty much solves “dictionary” problem. But what about substrings? • Relevance to DNA searches • idea: trie of all n2 substrings • equivalent to trie of all n suffixes. -

Balanced Trees Part One
Balanced Trees Part One Balanced Trees ● Balanced search trees are among the most useful and versatile data structures. ● Many programming languages ship with a balanced tree library. ● C++: std::map / std::set ● Java: TreeMap / TreeSet ● Many advanced data structures are layered on top of balanced trees. ● We’ll see several later in the quarter! Where We're Going ● B-Trees (Today) ● A simple type of balanced tree developed for block storage. ● Red/Black Trees (Today/Thursday) ● The canonical balanced binary search tree. ● Augmented Search Trees (Thursday) ● Adding extra information to balanced trees to supercharge the data structure. Outline for Today ● BST Review ● Refresher on basic BST concepts and runtimes. ● Overview of Red/Black Trees ● What we're building toward. ● B-Trees and 2-3-4 Trees ● Simple balanced trees, in depth. ● Intuiting Red/Black Trees ● A much better feel for red/black trees. A Quick BST Review Binary Search Trees ● A binary search tree is a binary tree with 9 the following properties: 5 13 ● Each node in the BST stores a key, and 1 6 10 14 optionally, some auxiliary information. 3 7 11 15 ● The key of every node in a BST is strictly greater than all keys 2 4 8 12 to its left and strictly smaller than all keys to its right. Binary Search Trees ● The height of a binary search tree is the 9 length of the longest path from the root to a 5 13 leaf, measured in the number of edges. 1 6 10 14 ● A tree with one node has height 0. -

Optimal Time and Space Construction of Suffix Arrays and LCP
Optimal Time and Space Construction of Suffix Arrays and LCP Arrays for Integer Alphabets Keisuke Goto Fujitsu Laboratories Ltd. Kawasaki, Japan [email protected] Abstract Suffix arrays and LCP arrays are one of the most fundamental data structures widely used for various kinds of string processing. We consider two problems for a read-only string of length N over an integer alphabet [1, . , σ] for 1 ≤ σ ≤ N, the string contains σ distinct characters, the construction of the suffix array, and a simultaneous construction of both the suffix array and LCP array. For the word RAM model, we propose algorithms to solve both of the problems in O(N) time by using O(1) extra words, which are optimal in time and space. Extra words means the required space except for the space of the input string and output suffix array and LCP array. Our contribution improves the previous most efficient algorithms, O(N) time using σ + O(1) extra words by [Nong, TOIS 2013] and O(N log N) time using O(1) extra words by [Franceschini and Muthukrishnan, ICALP 2007], for constructing suffix arrays, and it improves the previous most efficient solution that runs in O(N) time using σ + O(1) extra words for constructing both suffix arrays and LCP arrays through a combination of [Nong, TOIS 2013] and [Manzini, SWAT 2004]. 2012 ACM Subject Classification Theory of computation, Pattern matching Keywords and phrases Suffix Array, Longest Common Prefix Array, In-Place Algorithm Acknowledgements We wish to thank Takashi Kato, Shunsuke Inenaga, Hideo Bannai, Dominik Köppl, and anonymous reviewers for their many valuable suggestions on improving the quality of this paper, and especially, we also wish to thank an anonymous reviewer for giving us a simpler algorithm for computing LCP arrays as described in Section 5. -

Suffix Trees
JASS 2008 Trees - the ubiquitous structure in computer science and mathematics Suffix Trees Caroline L¨obhard St. Petersburg, 9.3. - 19.3. 2008 1 Contents 1 Introduction to Suffix Trees 3 1.1 Basics . 3 1.2 Getting a first feeling for the nice structure of suffix trees . 4 1.3 A historical overview of algorithms . 5 2 Ukkonen’s on-line space-economic linear-time algorithm 6 2.1 High-level description . 6 2.2 Using suffix links . 7 2.3 Edge-label compression and the skip/count trick . 8 2.4 Two more observations . 9 3 Generalised Suffix Trees 9 4 Applications of Suffix Trees 10 References 12 2 1 Introduction to Suffix Trees A suffix tree is a tree-like data-structure for strings, which affords fast algorithms to find all occurrences of substrings. A given String S is preprocessed in O(|S|) time. Afterwards, for any other string P , one can decide in O(|P |) time, whether P can be found in S and denounce all its exact positions in S. This linear worst case time bound depending only on the length of the (shorter) string |P | is special and important for suffix trees since an amount of applications of string processing has to deal with large strings S. 1.1 Basics In this paper, we will denote the fixed alphabet with Σ, single characters with lower-case letters x, y, ..., strings over Σ with upper-case or Greek letters P, S, ..., α, σ, τ, ..., Trees with script letters T , ... and inner nodes of trees (that is, all nodes despite of root and leaves) with lower-case letters u, v, ... -
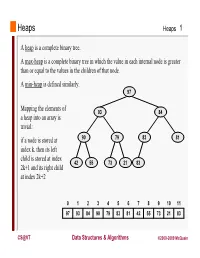
Heaps a Heap Is a Complete Binary Tree. a Max-Heap Is A
Heaps Heaps 1 A heap is a complete binary tree. A max-heap is a complete binary tree in which the value in each internal node is greater than or equal to the values in the children of that node. A min-heap is defined similarly. 97 Mapping the elements of 93 84 a heap into an array is trivial: if a node is stored at 90 79 83 81 index k, then its left child is stored at index 42 55 73 21 83 2k+1 and its right child at index 2k+2 01234567891011 97 93 84 90 79 83 81 42 55 73 21 83 CS@VT Data Structures & Algorithms ©2000-2009 McQuain Building a Heap Heaps 2 The fact that a heap is a complete binary tree allows it to be efficiently represented using a simple array. Given an array of N values, a heap containing those values can be built, in situ, by simply “sifting” each internal node down to its proper location: - start with the last 73 73 internal node * - swap the current 74 81 74 * 93 internal node with its larger child, if 79 90 93 79 90 81 necessary - then follow the swapped node down 73 * 93 - continue until all * internal nodes are 90 93 90 73 done 79 74 81 79 74 81 CS@VT Data Structures & Algorithms ©2000-2009 McQuain Heap Class Interface Heaps 3 We will consider a somewhat minimal maxheap class: public class BinaryHeap<T extends Comparable<? super T>> { private static final int DEFCAP = 10; // default array size private int size; // # elems in array private T [] elems; // array of elems public BinaryHeap() { . -

Full-Text and Keyword Indexes for String Searching
Technische Universität München Full-text and Keyword Indexes for String Searching by Aleksander Cisłak arXiv:1508.06610v1 [cs.DS] 26 Aug 2015 Master’s Thesis in Informatics Department of Informatics Technische Universität München Full-text and Keyword Indexes for String Searching Indizierung von Volltexten und Keywords für Textsuche Author: Aleksander Cisłak ([email protected]) Supervisor: prof. Burkhard Rost Advisors: prof. Szymon Grabowski; Tatyana Goldberg, MSc Master’s Thesis in Informatics Department of Informatics August 2015 Declaration of Authorship I, Aleksander Cisłak, confirm that this master’s thesis is my own work and I have docu- mented all sources and material used. Signed: Date: ii Abstract String searching consists in locating a substring in a longer text, and two strings can be approximately equal (various similarity measures such as the Hamming distance exist). Strings can be defined very broadly, and they usually contain natural language and biological data (DNA, proteins), but they can also represent other kinds of data such as music or images. One solution to string searching is to use online algorithms which do not preprocess the input text, however, this is often infeasible due to the massive sizes of modern data sets. Alternatively, one can build an index, i.e. a data structure which aims to speed up string matching queries. The indexes are divided into full-text ones which operate on the whole input text and can answer arbitrary queries and keyword indexes which store a dictionary of individual words. In this work, we present a literature review for both index categories as well as our contributions (which are mostly practice-oriented). -

CSCI 333 Data Structures Binary Trees Binary Tree Example Full And
Notes with the dark blue background CSCI 333 were prepared by the textbook author Data Structures Clifford A. Shaffer Chapter 5 Department of Computer Science 18, 20, 23, and 25 September 2002 Virginia Tech Copyright © 2000, 2001 Binary Trees Binary Tree Example A binary tree is made up of a finite set of Notation: Node, nodes that is either empty or consists of a children, edge, node called the root together with two parent, ancestor, binary trees, called the left and right descendant, path, subtrees, which are disjoint from each depth, height, level, other and from the root. leaf node, internal node, subtree. Full and Complete Binary Trees Full Binary Tree Theorem (1) Full binary tree: Each node is either a leaf or Theorem: The number of leaves in a non-empty internal node with exactly two non-empty children. full binary tree is one more than the number of internal nodes. Complete binary tree: If the height of the tree is d, then all leaves except possibly level d are Proof (by Mathematical Induction): completely full. The bottom level has all nodes to the left side. Base case: A full binary tree with 1 internal node must have two leaf nodes. Induction Hypothesis: Assume any full binary tree T containing n-1 internal nodes has n leaves. 1 Full Binary Tree Theorem (2) Full Binary Tree Corollary Induction Step: Given tree T with n internal Theorem: The number of null pointers in a nodes, pick internal node I with two leaf children. non-empty tree is one more than the Remove I’s children, call resulting tree T’. -

Search Trees
Lecture III Page 1 “Trees are the earth’s endless effort to speak to the listening heaven.” – Rabindranath Tagore, Fireflies, 1928 Alice was walking beside the White Knight in Looking Glass Land. ”You are sad.” the Knight said in an anxious tone: ”let me sing you a song to comfort you.” ”Is it very long?” Alice asked, for she had heard a good deal of poetry that day. ”It’s long.” said the Knight, ”but it’s very, very beautiful. Everybody that hears me sing it - either it brings tears to their eyes, or else -” ”Or else what?” said Alice, for the Knight had made a sudden pause. ”Or else it doesn’t, you know. The name of the song is called ’Haddocks’ Eyes.’” ”Oh, that’s the name of the song, is it?” Alice said, trying to feel interested. ”No, you don’t understand,” the Knight said, looking a little vexed. ”That’s what the name is called. The name really is ’The Aged, Aged Man.’” ”Then I ought to have said ’That’s what the song is called’?” Alice corrected herself. ”No you oughtn’t: that’s another thing. The song is called ’Ways and Means’ but that’s only what it’s called, you know!” ”Well, what is the song then?” said Alice, who was by this time completely bewildered. ”I was coming to that,” the Knight said. ”The song really is ’A-sitting On a Gate’: and the tune’s my own invention.” So saying, he stopped his horse and let the reins fall on its neck: then slowly beating time with one hand, and with a faint smile lighting up his gentle, foolish face, he began.. -

Tree Structures
Tree Structures Definitions: o A tree is a connected acyclic graph. o A disconnected acyclic graph is called a forest o A tree is a connected digraph with these properties: . There is exactly one node (Root) with in-degree=0 . All other nodes have in-degree=1 . A leaf is a node with out-degree=0 . There is exactly one path from the root to any leaf o The degree of a tree is the maximum out-degree of the nodes in the tree. o If (X,Y) is a path: X is an ancestor of Y, and Y is a descendant of X. Root X Y CSci 1112 – Algorithms and Data Structures, A. Bellaachia Page 1 Level of a node: Level 0 or 1 1 or 2 2 or 3 3 or 4 Height or depth: o The depth of a node is the number of edges from the root to the node. o The root node has depth zero o The height of a node is the number of edges from the node to the deepest leaf. o The height of a tree is a height of the root. o The height of the root is the height of the tree o Leaf nodes have height zero o A tree with only a single node (hence both a root and leaf) has depth and height zero. o An empty tree (tree with no nodes) has depth and height −1. o It is the maximum level of any node in the tree. CSci 1112 – Algorithms and Data Structures, A. -

Binary Search Tree
ADT Binary Search Tree! Ellen Walker! CPSC 201 Data Structures! Hiram College! Binary Search Tree! •" Value-based storage of information! –" Data is stored in order! –" Data can be retrieved by value efficiently! •" Is a binary tree! –" Everything in left subtree is < root! –" Everything in right subtree is >root! –" Both left and right subtrees are also BST#s! Operations on BST! •" Some can be inherited from binary tree! –" Constructor (for empty tree)! –" Inorder, Preorder, and Postorder traversal! •" Some must be defined ! –" Insert item! –" Delete item! –" Retrieve item! The Node<E> Class! •" Just as for a linked list, a node consists of a data part and links to successor nodes! •" The data part is a reference to type E! •" A binary tree node must have links to both its left and right subtrees! The BinaryTree<E> Class! The BinaryTree<E> Class (continued)! Overview of a Binary Search Tree! •" Binary search tree definition! –" A set of nodes T is a binary search tree if either of the following is true! •" T is empty! •" Its root has two subtrees such that each is a binary search tree and the value in the root is greater than all values of the left subtree but less than all values in the right subtree! Overview of a Binary Search Tree (continued)! Searching a Binary Tree! Class TreeSet and Interface Search Tree! BinarySearchTree Class! BST Algorithms! •" Search! •" Insert! •" Delete! •" Print values in order! –" We already know this, it#s inorder traversal! –" That#s why it#s called “in order”! Searching the Binary Tree! •" If the tree is