(12) Patent Application Publication (10) Pub. No.: US 2010/0267582 A1 BARD Et Al
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Effect of a Caloric Restriction Based On
UNIVERSIDAD AUTÓNOMA DE MADRID FACULTAD DE CIENCIAS DEPARTAMENTO DE BIOLOGÍA DOCTORAL THESIS Biology PhD “Effect of a caloric restriction based on the Mediterranean diet and intake of traditional Mediterranean foods on the expression of microRNAs regulating molecular processes associated with aging” INSTITUTO MADRILEÑO DE ESTUDIOS AVANZADOS EN ALIMENTACIÓN (IMDEA FOOD INSTITUTE) VÍCTOR MICÓ MORENO Madrid, 2018 UNIVERSIDAD AUTÓNOMA DE MADRID FACULTAD DE CIENCIAS DEPARTAMENTO DE BIOLOGÍA DOCTORAL THESIS Biology PhD “Effect of a caloric restriction based on the Mediterranean diet and intake of traditional Mediterranean foods on the expression of microRNAs regulating molecular processes associated with aging” INSTITUTO MADRILEÑO DE ESTUDIOS AVANZADOS EN ALIMENTACIÓN (IMDEA FOOD INSTITUTE) Memoria presentada por: Víctor Micó Moreno Para optar al grado de: DOCTOR EN BIOLOGÍA Doña Lidia Ángeles Daimiel Ruíz, Doctora en Biología Celular y Genética por la Universidad Autónoma de Madrid, investigadora del Instituto IMDEA Alimentación, informa favorablemente la solicitud de autorización de defensa de la tesis doctoral con el Título: “Effect of a caloric restriction based on the Mediterranean diet and intake of traditional Mediterranean foods on the expression of microRNAs regulating molecular processes associated with aging”, presentada por Don Víctor Micó Moreno para optar al grado de Doctor en Biología. Este trabajo ha sido realizado en el Instituto Madrileño de Estudios Avanzados en Alimentación (IMDEA Alimentación) bajo su dirección, y cumple satisfactoriamente las condiciones requeridas por el Departamento de Biología de la Universidad Autónoma de Madrid para optar al Título de Doctor. Ha actuado como tutor académico, y presenta su conformidad el Dr. Carlos Francisco Sentís Castaño, vicedecano de Personal Docente e Investigador y profesor titular del Departamento de Biología de la Facultad de Ciencias de la Universidad Autónoma de Madrid. -

Figure S1. HAEC ROS Production and ML090 NOX5-Inhibition
Figure S1. HAEC ROS production and ML090 NOX5-inhibition. (a) Extracellular H2O2 production in HAEC treated with ML090 at different concentrations and 24 h after being infected with GFP and NOX5-β adenoviruses (MOI 100). **p< 0.01, and ****p< 0.0001 vs control NOX5-β-infected cells (ML090, 0 nM). Results expressed as mean ± SEM. Fold increase vs GFP-infected cells with 0 nM of ML090. n= 6. (b) NOX5-β overexpression and DHE oxidation in HAEC. Representative images from three experiments are shown. Intracellular superoxide anion production of HAEC 24 h after infection with GFP and NOX5-β adenoviruses at different MOIs treated or not with ML090 (10 nM). MOI: Multiplicity of infection. Figure S2. Ontology analysis of HAEC infected with NOX5-β. Ontology analysis shows that the response to unfolded protein is the most relevant. Figure S3. UPR mRNA expression in heart of infarcted transgenic mice. n= 12-13. Results expressed as mean ± SEM. Table S1: Altered gene expression due to NOX5-β expression at 12 h (bold, highlighted in yellow). N12hvsG12h N18hvsG18h N24hvsG24h GeneName GeneDescription TranscriptID logFC p-value logFC p-value logFC p-value family with sequence similarity NM_052966 1.45 1.20E-17 2.44 3.27E-19 2.96 6.24E-21 FAM129A 129. member A DnaJ (Hsp40) homolog. NM_001130182 2.19 9.83E-20 2.94 2.90E-19 3.01 1.68E-19 DNAJA4 subfamily A. member 4 phorbol-12-myristate-13-acetate- NM_021127 0.93 1.84E-12 2.41 1.32E-17 2.69 1.43E-18 PMAIP1 induced protein 1 E2F7 E2F transcription factor 7 NM_203394 0.71 8.35E-11 2.20 2.21E-17 2.48 1.84E-18 DnaJ (Hsp40) homolog. -

1 Novel Expression Signatures Identified by Transcriptional Analysis
ARD Online First, published on October 7, 2009 as 10.1136/ard.2009.108043 Ann Rheum Dis: first published as 10.1136/ard.2009.108043 on 7 October 2009. Downloaded from Novel expression signatures identified by transcriptional analysis of separated leukocyte subsets in SLE and vasculitis 1Paul A Lyons, 1Eoin F McKinney, 1Tim F Rayner, 1Alexander Hatton, 1Hayley B Woffendin, 1Maria Koukoulaki, 2Thomas C Freeman, 1David RW Jayne, 1Afzal N Chaudhry, and 1Kenneth GC Smith. 1Cambridge Institute for Medical Research and Department of Medicine, Addenbrooke’s Hospital, Hills Road, Cambridge, CB2 0XY, UK 2Roslin Institute, University of Edinburgh, Roslin, Midlothian, EH25 9PS, UK Correspondence should be addressed to Dr Paul Lyons or Prof Kenneth Smith, Department of Medicine, Cambridge Institute for Medical Research, Addenbrooke’s Hospital, Hills Road, Cambridge, CB2 0XY, UK. Telephone: +44 1223 762642, Fax: +44 1223 762640, E-mail: [email protected] or [email protected] Key words: Gene expression, autoimmune disease, SLE, vasculitis Word count: 2,906 The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non-exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in Annals of the Rheumatic Diseases and any other BMJPGL products to exploit all subsidiary rights, as set out in their licence (http://ard.bmj.com/ifora/licence.pdf). http://ard.bmj.com/ on September 29, 2021 by guest. Protected copyright. 1 Copyright Article author (or their employer) 2009. -

2U11/13U624 A2
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date ft i 20 October 2011 (20.10.2011) 2U11/13U624 A2 (51) International Patent Classification: AO, AT, AU, AZ, BA, BB, BG, BH, BR, BW, BY, BZ, C12N 5/10 (2006.01) CI2N 5/074 (2010.01) CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, C12N 15/113 (2010.01) C07H 21/00 (2006.01) DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, (21) International Application Number: KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, PCT/US201 1/032679 ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, (22) International Filing Date: NO, NZ, OM, PE, PG, PH, PL, PT, RO, RS, RU, SC, SD, 15 April 201 1 (15.04.201 1) SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every (26) Publication Language: English kind of regional protection available): ARIPO (BW, GH, (30) Priority Data: GM, KE, LR, LS, MW, MZ, NA, SD, SL, SZ, TZ, UG, 61/325,003 16 April 2010 (16.04.2010) US ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, MD, RU, TJ, 61/387,220 28 September 2010 (28.09.2010) US TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, ΓΓ, LT, LU, (71) Applicant (for all designated States except US): IM¬ LV, MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, MUNE DISEASE INSTITUTE, INC. -

BMC Genomics Biomed Central
BMC Genomics BioMed Central Research article Open Access Genome-wide changes accompanying knockdown of fatty acid synthase in breast cancer Lynn M Knowles and Jeffrey W Smith* Address: Cancer Research Center, Burnham Institute for Medical Research, La Jolla, CA 92037, USA Email: Lynn M Knowles - [email protected]; Jeffrey W Smith* - [email protected] * Corresponding author Published: 12 June 2007 Received: 8 November 2006 Accepted: 12 June 2007 BMC Genomics 2007, 8:168 doi:10.1186/1471-2164-8-168 This article is available from: http://www.biomedcentral.com/1471-2164/8/168 © 2007 Knowles and Smith; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: The lipogenic enzyme fatty acid synthase (FAS) is up-regulated in a wide variety of cancers, and is considered a potential metabolic oncogene by virtue of its ability to enhance tumor cell survival. Inhibition of tumor FAS causes both cell cycle arrest and apoptosis, indicating FAS is a promising target for cancer treatment. Results: Here, we used gene expression profiling to conduct a global study of the cellular processes affected by siRNA mediated knockdown of FAS in MDA-MB-435 mammary carcinoma cells. The study identified 169 up-regulated genes (≥ 1.5 fold) and 110 down-regulated genes (≤ 0.67 fold) in response to knockdown of FAS. These genes regulate several aspects of tumor function, including metabolism, cell survival/proliferation, DNA replication/transcription, and protein degradation. -

1 Novel Expression Signatures Identified by Transcriptional Analysis
ARD Online First, published on October 8, 2009 as 10.1136/ard.2009.108043 Ann Rheum Dis: first published as 10.1136/ard.2009.108043 on 7 October 2009. Downloaded from Novel expression signatures identified by transcriptional analysis of separated leukocyte subsets in SLE and vasculitis 1Paul A Lyons, 1Eoin F McKinney, 1Tim F Rayner, 1Alexander Hatton, 1Hayley B Woffendin, 1Maria Koukoulaki, 2Thomas C Freeman, 1David RW Jayne, 1Afzal N Chaudhry, and 1Kenneth GC Smith. 1Cambridge Institute for Medical Research and Department of Medicine, Addenbrooke’s Hospital, Hills Road, Cambridge, CB2 0XY, UK 2Roslin Institute, University of Edinburgh, Roslin, Midlothian, EH25 9PS, UK Correspondence should be addressed to Dr Paul Lyons or Prof Kenneth Smith, Department of Medicine, Cambridge Institute for Medical Research, Addenbrooke’s Hospital, Hills Road, Cambridge, CB2 0XY, UK. Telephone: +44 1223 762642, Fax: +44 1223 762640, E-mail: [email protected] or [email protected] Key words: Gene expression, autoimmune disease, SLE, vasculitis Word count: 2,906 The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non-exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in Annals of the Rheumatic Diseases and any other BMJPGL products to exploit all subsidiary rights, as set out in their licence (http://ard.bmj.com/ifora/licence.pdf). http://ard.bmj.com/ on October 2, 2021 by guest. Protected copyright. 1 Copyright Article author (or their employer) 2009. -

Post-Transcriptional Regulation of Gene Expression in Response to Iron Deficiency in Saccharomyces Cerevisiae by Sandra Viviana
Post-transcriptional Regulation of Gene Expression in Response to Iron Deficiency in Saccharomyces cerevisiae by Sandra Viviana Vergara Program in Genetics and Genomics Duke University Date:_______________________ Approved: ___________________________ Dennis J. Thiele, Ph.D., Supervisor ___________________________ Matthias Gromeier, M.D. ___________________________ Jack Keene, Ph.D. ___________________________ Daniel Lew, Ph.D. ___________________________ William Marzluff, Ph.D. Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Program in Genetics and Genomics of Duke University 2010 ABSTRACT Post-transcriptional Regulation of Gene Expression in Response to Iron Deficiency in Saccharomyces cerevisiae by Sandra Viviana Vergara Program in Genetics and Genomics Duke University Date:_______________________ Approved: ___________________________ Dennis J. Thiele, Ph.D., Supervisor ___________________________ Matthias Gromeier, M.D. ___________________________ Jack Keene, Ph.D. ___________________________ Daniel Lew, Ph.D. ___________________________ William Marzluff, Ph.D. An abstract of a dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Program in Genetics and Genomics in the Graduate School of Duke University 2010 Copyright by Sandra Viviana Vergara 2010 Abstract The ability of iron (Fe) to easily transition between two valence states makes it a preferred co-factor for innumerable biochemical reactions, ranging from cellular energy production, to oxygen transport, to DNA synthesis and chromatin modification. While Fe is highly abundant on the crust of the earth, its insolubility at neutral pH limits its bioavailability. As a consequence, organisms have evolved sophisticated mechanisms of adaptation to conditions of scarce Fe availability. Studies in the baker’s yeast Saccharomyces cerevisiae have shed light into the cellular mechanisms by which cells respond to limited Fe-availability. -

Allicin Reverses Diabetes-Induced Dysfunction of Human Coronary Artery Endothelial Cells
Philadelphia College of Osteopathic Medicine DigitalCommons@PCOM PCOM Biomedical Studies Student Scholarship Student Dissertations, Theses and Papers 5-2019 Allicin Reverses Diabetes-Induced Dysfunction of Human Coronary Artery Endothelial Cells Daniel Horuzsko Follow this and additional works at: https://digitalcommons.pcom.edu/biomed Part of the Medicine and Health Sciences Commons Recommended Citation Horuzsko, Daniel, "Allicin Reverses Diabetes-Induced Dysfunction of Human Coronary Artery Endothelial Cells" (2019). PCOM Biomedical Studies Student Scholarship. 187. https://digitalcommons.pcom.edu/biomed/187 This Thesis is brought to you for free and open access by the Student Dissertations, Theses and Papers at DigitalCommons@PCOM. It has been accepted for inclusion in PCOM Biomedical Studies Student Scholarship by an authorized administrator of DigitalCommons@PCOM. For more information, please contact [email protected]. Philadelphia College of Osteopathic Medicine Biomedical Sciences Allicin Reverses Diabetes-Induced Dysfunction of Human Coronary Artery Endothelial Cells A Thesis in Biomedical Science by Daniel Horuzsko Copyright May 2019 Daniel Horuzsko Biomedical Sciences Degree Program Thesis Signatory Page We approve the thesis of Daniel Horuzsko. Richard E. White, Ph.D., F.A.H.A. Date of Signature Professor of Neuroscience, Pharmacology, and Physiology Associate Director of Biomedical Sciences Program Philadelphia College of Osteopathic Medicine-Georgia Thesis Advisor Shu Zhu, M.D., Ph.D. Date of Signature Associate Professor of Neuroscience, Pharmacology, and Physiology Department of Biomedical Sciences Philadelphia College of Osteopathic Medicine-Georgia Committee Member Shafik Habal, M.D. Date of Signature Associate Professor of Microbiology and Immunology Department of Biomedical Sciences Philadelphia College of Osteopathic Medicine-Georgia Committee Member Richard E. White, Ph.D., F.A.H.A. -

Mechanistic Studies of Guanine-Rich Oligonucleotides
University of Louisville ThinkIR: The University of Louisville's Institutional Repository Electronic Theses and Dissertations 5-2006 Mechanistic studies of guanine-rich oligonucleotides. Allicia C. Girvan 1978- University of Louisville Follow this and additional works at: https://ir.library.louisville.edu/etd Recommended Citation Girvan, Allicia C. 1978-, "Mechanistic studies of guanine-rich oligonucleotides." (2006). Electronic Theses and Dissertations. Paper 501. https://doi.org/10.18297/etd/501 This Doctoral Dissertation is brought to you for free and open access by ThinkIR: The University of Louisville's Institutional Repository. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of ThinkIR: The University of Louisville's Institutional Repository. This title appears here courtesy of the author, who has retained all other copyrights. For more information, please contact [email protected]. MECHANISTIC STUDIES OF GUANINE-RICH OLIGONUCLEOTIDES By Allicia C. Girvan B. S., Southwest Baptist University, 2000 A Dissertation Submitted to the Faculty of the Graduate School of the University of Louisville in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Department of Biochemistry and Molecular Biology University of Louisville Louisville, Kentucky May 2006 MECHANISTIC STUDIES OF GUANINE-RICH OLIGONUCLEOTIDES By Allicia C. Girvan B.S., Southwest Baptist University A Dissertation Approved on April 28, 2006 By the following Dissertation Committee: Dissertation Director ii DEDICATION To my loving husband Sonny Thank you for your unconditional love and encouragement everyday 111 ACKNOWLEDGEMENTS I hold such admiration and respect for my mentor Dr. Paula Bates. Her guidance and training has provided me with the foundation in which I will use throughout my entire career. -

Supplementary Table S4
Supplementary Table S4 - Genes downregulated by anti-MYCN PNA ProbeID Symbol Description RH30ctrl_SignalRH30mut__SignalRH30pna_Signalpna vs mut pna vs ctrl 210842_at NRP2 neuropilin 2 109.8 83.2 3.4 -4.613 -5.013 219239_s_at ZNF654 zinc finger protein 654 155.9 140.8 6.8 -4.372 -4.519 220786_s_at SLC38A4 solute carrier family 38, member 4 91.8 135.1 17.7 -2.932 -2.375 206205_at MPHOSPH9 M-phase phosphoprotein 9 208.2 235.7 39.2 -2.588 -2.409 201171_at ATP6V0E ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e 93.5 105 19.1 -2.459 -2.291 204732_s_at TRIM23 tripartite motif-containing 23 132.9 126.3 31 -2.027 -2.100 221079_s_at METTL2B methyltransferase like 2B 141.8 236.9 58.8 -2.010 -1.270 216971_s_at PLEC1 plectin 1, intermediate filament binding protein 500kDa 170 176.3 44.7 -1.980 -1.927 221703_at BRIP1 BRCA1 interacting protein C-terminal helicase 1 137 200.4 52.6 -1.930 -1.381 213647_at DNA2L DNA2 DNA replication helicase 2-like (yeast) 180.5 226.1 61.4 -1.881 -1.556 217547_x_at ZNF675 zinc finger protein 675 89.6 93.3 26 -1.843 -1.785 209865_at SLC35A3 solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 191.6 171.3 50.4 -1.765 -1.927 AFFX-M27830_5_atSOX18 SRY (sex determining region Y)-box 18 755.4 982.7 295.8 -1.732 -1.353 212177_at C6orf111 chromosome 6 open reading frame 111 453.6 484.7 147.8 -1.713 -1.618 211088_s_at PLK4 polo-like kinase 4 (Drosophila) 174.6 122.1 37.4 -1.707 -2.223 213880_at LGR5 leucine-rich repeat-containing G protein-coupled receptor 5 172 178 56.2 -1.663 -1.614 201534_s_at -

4 353 Skin Oral 1 B
A B Supplementary Figure S1: Differentially expressed piRNAs during skin and oral mucosal wound healing. (A) piRNA 0hr-1 0hr-3 0hr-2 24hr-2 24hr-1 24hr-3 5day-3 5day-1 5day-2 0hr-1 0hr-2 0hr-3 24hr-2 24hr-1 24hr-3 5day-1 5day-2 5day-3 profiles were obtained on mouse skin and oral mucosal (palate) wound healing time course (0hr, 24 hr, and 5 day). A total of 357 differentially expressed piRNA were identified during skin wound healing (Bonferroni adjusted P value <0.05). See Supplementary Table 1A for the full list. C (B) Five differentially expressed piRNA skin were identified during oral mucosal wound healing (P value <0.01, list presented in Supplementary Table 1B). Note: more 353 stringent statistical cut-off ((Bonferroni adjusted P value) yield 0 differentially expressed piRNA gene. (C) Venn diagram illustrates overlaps between differentially 4 expressedpiRNAsinskinandoral oral 1 mucosal wound healing. min max Supplementary Table S1a: Differentially expressed piRNAs in skin wound healing Mean StDev piRNA 0 hr 24 hr 5 day 0 hr 24 hr 5 day pVal adj P piR‐mmu‐15927330 5.418351 11.39746 10.799 0.34576 0.253492 0.154802 6.32E‐14 6.97E‐11 piR‐mmu‐49559417 5.301647 10.10777 9.816878 0.441719 0.222335 0.032479 3.95E‐12 4.36E‐09 piR‐mmu‐30053093 6.32531 11.26384 4.020902 0.280841 1.057847 0.178798 5.21E‐12 5.76E‐09 piR‐mmu‐29303577 5.15005 10.47662 9.52175 0.554877 0.26622 0.163283 1.53E‐11 1.69E‐08 piR‐mmu‐49254706 5.187673 10.19644 9.622374 0.520671 0.330378 0.191116 1.96E‐11 2.16E‐08 piR‐mmu‐49005170 5.415133 9.639725 9.565967 0.411507 0.281143 -
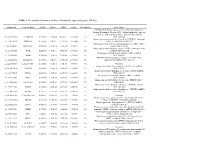
Table 1 the Statistical Metrics for Key Differentially Expressed Genes (Degs)
Table 1 The statistical metrics for key differentially expressed genes (DEGs) Agiliant Id Gene Symbol logFC pValue FDR tvalue Regulation Gene Name PREDICTED: Homo sapiens similar to Filamin-C (Gamma- filamin) (Filamin-2) (Protein FLNc) (Actin-binding-like protein) (ABP-L) (ABP-280-like protein) (LOC649056), mRNA A_24_P237896 LOC649056 0.509843 9.18E-14 4.54E-11 12.07302 Up [XR_018580] Homo sapiens programmed cell death 10 (PDCD10), transcript A_23_P18325 PDCD10 0.243111 2.8E-12 9.24E-10 10.62808 Up variant 1, mRNA [NM_007217] Homo sapiens Rho GTPase activating protein 27 (ARHGAP27), A_23_P141335 ARHGAP27 0.492709 3.97E-12 1.22E-09 10.48571 Up mRNA [NM_199282] Homo sapiens tubby homolog (mouse) (TUB), transcript variant A_23_P53110 TUB 0.528219 1.77E-11 4.56E-09 9.891033 Up 1, mRNA [NM_003320] Homo sapiens MyoD family inhibitor (MDFI), mRNA A_23_P42168 MDFI 0.314474 1.81E-10 3.74E-08 8.998697 Up [NM_005586] PREDICTED: Homo sapiens hypothetical LOC644701 A_32_P56890 LOC644701 0.444703 3.6E-10 7.09E-08 8.743973 Up (LOC644701), mRNA [XM_932316] A_32_P167111 A_32_P167111 0.873588 7.41E-10 1.4E-07 8.47781 Up Unknown Homo sapiens zinc finger protein 784 (ZNF784), mRNA A_24_P221424 ZNF784 0.686781 9.18E-10 1.68E-07 8.399687 Up [NM_203374] Homo sapiens lin-28 homolog (C. elegans) (LIN28), mRNA A_23_P74895 LIN28 0.218876 1.27E-09 2.24E-07 8.282224 Up [NM_024674] Homo sapiens ribosomal protein L5 (RPL5), mRNA A_23_P12140 RPL5 0.247598 1.81E-09 3.11E-07 8.154317 Up [NM_000969] Homo sapiens cDNA FLJ43841 fis, clone TESTI4006137.