Affymetrix Probe Set Gene Symbol
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Wiskott-Aldrich Syndrome: the Actin Cytoskeleton and Immune Cell Function
Disease Markers 29 (2010) 157–175 157 DOI 10.3233/DMA-2010-0735 IOS Press The Wiskott-Aldrich syndrome: The actin cytoskeleton and immune cell function Michael P. Blundella, Austen Wortha,b, Gerben Boumaa and Adrian J. Thrashera,b,∗ aMolecular Immunology Unit, UCL Institute of Child Health, London, UK bDepartment of Immunology, Great Ormond Street Hospital NHS Trust, Great Ormond Street, London, UK Abstract. Wiskott-Aldrich syndrome (WAS) is a rare X-linked recessive primary immunodeficiency characterised by immune dysregulation, microthrombocytopaenia, eczema and lymphoid malignancies. Mutations in the WAS gene can lead to distinct syndrome variations which largely, although not exclusively, depend upon the mutation. Premature termination and deletions abrogate Wiskott-Aldrich syndrome protein (WASp) expression and lead to severe disease (WAS). Missense mutations usually result in reduced protein expression and the phenotypically milder X-linked thrombocytopenia (XLT) or attenuated WAS [1–3]. More recently however novel activating mutations have been described that give rise to X-linked neutropenia (XLN), a third syndrome defined by neutropenia with variable myelodysplasia [4–6]. WASP is key in transducing signals from the cell surface to the actin cytoskeleton, and a lack of WASp results in cytoskeletal defects that compromise multiple aspects of normal cellular activity including proliferation, phagocytosis, immune synapse formation, adhesion and directed migration. Keywords: Wiskott-Aldrich syndrome, actin polymerization, lymphocytes, -

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Functional Roles of Bromodomain Proteins in Cancer
cancers Review Functional Roles of Bromodomain Proteins in Cancer Samuel P. Boyson 1,2, Cong Gao 3, Kathleen Quinn 2,3, Joseph Boyd 3, Hana Paculova 3 , Seth Frietze 3,4,* and Karen C. Glass 1,2,4,* 1 Department of Pharmaceutical Sciences, Albany College of Pharmacy and Health Sciences, Colchester, VT 05446, USA; [email protected] 2 Department of Pharmacology, Larner College of Medicine, University of Vermont, Burlington, VT 05405, USA; [email protected] 3 Department of Biomedical and Health Sciences, University of Vermont, Burlington, VT 05405, USA; [email protected] (C.G.); [email protected] (J.B.); [email protected] (H.P.) 4 University of Vermont Cancer Center, Burlington, VT 05405, USA * Correspondence: [email protected] (S.F.); [email protected] (K.C.G.) Simple Summary: This review provides an in depth analysis of the role of bromodomain-containing proteins in cancer development. As readers of acetylated lysine on nucleosomal histones, bromod- omain proteins are poised to activate gene expression, and often promote cancer progression. We examined changes in gene expression patterns that are observed in bromodomain-containing proteins and associated with specific cancer types. We also mapped the protein–protein interaction network for the human bromodomain-containing proteins, discuss the cellular roles of these epigenetic regu- lators as part of nine different functional groups, and identify bromodomain-specific mechanisms in cancer development. Lastly, we summarize emerging strategies to target bromodomain proteins in cancer therapy, including those that may be essential for overcoming resistance. Overall, this review provides a timely discussion of the different mechanisms of bromodomain-containing pro- Citation: Boyson, S.P.; Gao, C.; teins in cancer, and an updated assessment of their utility as a therapeutic target for a variety of Quinn, K.; Boyd, J.; Paculova, H.; cancer subtypes. -
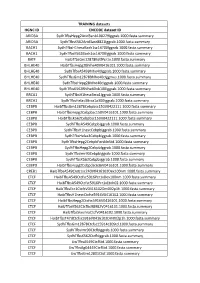
TRAINING Datasets HGNC ID ENCODE Dataset ID ARID3A
TRAINING datasets HGNC ID ENCODE dataset ID ARID3A SydhT+sHepg2Arid3anb100279Iggrab.1000.fasta.summary ARID3A SydhT+sK562Arid3asC8821Iggrab.1000.fasta.summary BACH1 SydhT+sH1hesCBaCh1sC14700Iggrab.1000.fasta.summary BACH1 SydhT+sK562BaCh1sC14700Iggrab.1000.fasta.summary BATF HaibT+sGm12878BaJPCr1x.1000.fasta.summary BHLHE40 HaibT+sHepg2Bhlhe40V0416101.1000.fasta.summary BHLHE40 SydhT+sA549Bhlhe40Iggrab.1000.fasta.summary BHLHE40 SydhT+sGm12878Bhlhe40CIggmus.1000.fasta.summary BHLHE40 SydhT+sHepg2Bhlhe40CIggrab.1000.fasta.summary BHLHE40 SydhT+sK562Bhlhe40nb100Iggrab.1000.fasta.summary BRCA1 SydhT+sH1hesCBrCa1Iggrab.1000.fasta.summary BRCA1 SydhT+sHelas3BrCa1a300Iggrab.1000.fasta.summary CEBPB HaibT+sGm12878CebpbsC150V0422111.1000.fasta.summary CEBPB HaibT+sHepg2CebpbsC150V0416101.1000.fasta.summary CEBPB HaibT+sK562CebpbsC150V0422111.1000.fasta.summary CEBPB SydhT+sA549CebpbIggrab.1000.fasta.summary CEBPB SydhT+sH1hesCCebpbIggrab.1000.fasta.summary CEBPB SydhT+sHelas3CebpbIggrab.1000.fasta.summary CEBPB SydhT+sHepg2CebpbForsklnStd.1000.fasta.summary CEBPB SydhT+sHepg2CebpbIggrab.1000.fasta.summary CEBPB SydhT+sImr90CebpbIggrab.1000.fasta.summary CEBPB SydhT+sK562CebpbIggrab.1000.fasta.summary CEBPD HaibT+sHepg2CebpdsC636V0416101.1000.fasta.summary CREB1 HaibT+sA549Creb1sC240V0416102Dex100nm.1000.fasta.summary CTCF HaibT+sA549CtCfsC5916PCr1xDex100nm.1000.fasta.summary CTCF HaibT+sA549CtCfsC5916PCr1xEtoh02.1000.fasta.summary CTCF HaibT+sECC1CtCfCV0416102Dm002p1h.1000.fasta.summary CTCF HaibT+sH1hesCCtCfsC5916V0416102.1000.fasta.summary -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Plasma Membrane Ca2+–Atpase in Rat and Human Odontoblasts Mediates Dentin Mineralization
biomolecules Article Plasma Membrane Ca2+–ATPase in Rat and Human Odontoblasts Mediates Dentin Mineralization Maki Kimura 1,†, Hiroyuki Mochizuki 1,†, Ryouichi Satou 2, Miyu Iwasaki 2, Eitoyo Kokubu 3, Kyosuke Kono 1, Sachie Nomura 1, Takeshi Sakurai 1, Hidetaka Kuroda 1,4,† and Yoshiyuki Shibukawa 1,*,† 1 Department of Physiology, Tokyo Dental College, 2-9-18, Kanda-Misaki-cho, Chiyoda-ku, Tokyo 101-0061, Japan; [email protected] (M.K.); [email protected] (H.M.); [email protected] (K.K.); [email protected] (S.N.); [email protected] (T.S.); [email protected] (H.K.) 2 Department of Epidemiology and Public Health, Tokyo Dental College, Chiyodaku, Tokyo 101-0061, Japan; [email protected] (R.S.); [email protected] (M.I.) 3 Department of Microbiology, Tokyo Dental College, Chiyodaku, Tokyo 101-0061, Japan; [email protected] 4 Department of Dental Anesthesiology, Kanagawa Dental University, 1-23, Ogawacho, Kanagawa, Yokosuka-shi 238-8570, Japan * Correspondence: [email protected] † These authors contributed equally to this study. Abstract: Intracellular Ca2+ signaling engendered by Ca2+ influx and mobilization in odontoblasts is critical for dentinogenesis induced by multiple stimuli at the dentin surface. Increased Ca2+ is exported by the Na+–Ca2+ exchanger (NCX) and plasma membrane Ca2+–ATPase (PMCA) to Citation: Kimura, M.; Mochizuki, H.; maintain Ca2+ homeostasis. We previously demonstrated a functional coupling between Ca2+ Satou, R.; Iwasaki, M.; Kokubu, E.; extrusion by NCX and its influx through transient receptor potential channels in odontoblasts. Kono, K.; Nomura, S.; Sakurai, T.; Although the presence of PMCA in odontoblasts has been previously described, steady-state levels of Kuroda, H.; Shibukawa, Y. -

Table 2. Significant
Table 2. Significant (Q < 0.05 and |d | > 0.5) transcripts from the meta-analysis Gene Chr Mb Gene Name Affy ProbeSet cDNA_IDs d HAP/LAP d HAP/LAP d d IS Average d Ztest P values Q-value Symbol ID (study #5) 1 2 STS B2m 2 122 beta-2 microglobulin 1452428_a_at AI848245 1.75334941 4 3.2 4 3.2316485 1.07398E-09 5.69E-08 Man2b1 8 84.4 mannosidase 2, alpha B1 1416340_a_at H4049B01 3.75722111 3.87309653 2.1 1.6 2.84852656 5.32443E-07 1.58E-05 1110032A03Rik 9 50.9 RIKEN cDNA 1110032A03 gene 1417211_a_at H4035E05 4 1.66015788 4 1.7 2.82772795 2.94266E-05 0.000527 NA 9 48.5 --- 1456111_at 3.43701477 1.85785922 4 2 2.8237185 9.97969E-08 3.48E-06 Scn4b 9 45.3 Sodium channel, type IV, beta 1434008_at AI844796 3.79536664 1.63774235 3.3 2.3 2.75319499 1.48057E-08 6.21E-07 polypeptide Gadd45gip1 8 84.1 RIKEN cDNA 2310040G17 gene 1417619_at 4 3.38875643 1.4 2 2.69163229 8.84279E-06 0.0001904 BC056474 15 12.1 Mus musculus cDNA clone 1424117_at H3030A06 3.95752801 2.42838452 1.9 2.2 2.62132809 1.3344E-08 5.66E-07 MGC:67360 IMAGE:6823629, complete cds NA 4 153 guanine nucleotide binding protein, 1454696_at -3.46081884 -4 -1.3 -1.6 -2.6026947 8.58458E-05 0.0012617 beta 1 Gnb1 4 153 guanine nucleotide binding protein, 1417432_a_at H3094D02 -3.13334396 -4 -1.6 -1.7 -2.5946297 1.04542E-05 0.0002202 beta 1 Gadd45gip1 8 84.1 RAD23a homolog (S. -

Hras Intracellular Trafficking and Signal Transduction Jodi Ho-Jung Mckay Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 2007 HRas intracellular trafficking and signal transduction Jodi Ho-Jung McKay Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Biological Phenomena, Cell Phenomena, and Immunity Commons, Cancer Biology Commons, Cell Biology Commons, Genetics and Genomics Commons, and the Medical Cell Biology Commons Recommended Citation McKay, Jodi Ho-Jung, "HRas intracellular trafficking and signal transduction" (2007). Retrospective Theses and Dissertations. 13946. https://lib.dr.iastate.edu/rtd/13946 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. HRas intracellular trafficking and signal transduction by Jodi Ho-Jung McKay A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Genetics Program of Study Committee: Janice E. Buss, Co-major Professor Linda Ambrosio, Co-major Professor Diane Bassham Drena Dobbs Ted Huiatt Iowa State University Ames, Iowa 2007 Copyright © Jodi Ho-Jung McKay, 2007. All rights reserved. UMI Number: 3274881 Copyright 2007 by McKay, Jodi Ho-Jung All rights reserved. UMI Microform 3274881 Copyright 2008 by ProQuest Information and Learning Company. All rights reserved. This microform edition is protected against unauthorized copying under Title 17, United States Code. ProQuest Information and Learning Company 300 North Zeeb Road P.O. -

Supplemental Information IRF4 Transcription Factor-Dependent Cd11b+ Dendritic Cells in Human and Mouse Control Mucosal IL-17 C
Immunity, Volume 38 Supplemental Information IRF4 Transcription Factor-Dependent CD11b+ Dendritic Cells in Human and Mouse Control Mucosal IL-17 Cytokine Responses Andreas Schlitzer, Naomi McGovern, Pearline Teo, Teresa Zelante, Koji Atarashi, Donovan Low, Adrian W.S. Ho, Peter See, Amanda Shin, Pavandip Singh Wasan, Guillaume Hoeffel, Benoit Malleret, Alexander Heiseke, Samantha Chew, Laura Jardine, Harriet A. Purvis, Catharien M.U. Hilkens, John Tam, Michael Poidinger, E. Richard Stanley, Anne B. K rug, Laurent Renia, Baalasubramanian Sivasankar, Lai Guan Ng, Matthew Collin, Paola Ricciardi-Castagnoli, Kenya Honda, Muzlifah Haniffa, and Florent Ginhoux Supplemental Inventory 1. Supplemental Figures and Tables Figure S1, Related to Figure 1 Figure S2, Related to Figure 2 and 3 Figure S3, Related to Figure 4 Figure S4, Related to Figure 5 Figure S5, Related to Figure 5 Figure S6, Related to Figure 7 Table S1, Related to Figure 3 Table S2, Related to Figure 6 Table S3, Related to Figure 6 2. Supplemental Experimental Procedures 3. Supplemental References Supplementary figure 1 Sorting strategy for mouse lung and small intestinal DC A Sorting strategy, Lung Singlets Dapi-CD45+ 250K 250K 250K 105 200K 200K 200K 104 A I - 150K 150K 150K C P C C 3 10 S A S S 100K 100K S 100K D S S 102 50K 50K 50K 0 0 0 0 0 50K 100K 150K 200K 250K 0 50K 100K 150K 200K 250K 0 103 104 105 0 103 104 105 FSC FSC-W CD45 GR1 Auto Fluor.- MHCII+ GR1- SSClow CD11c+ CD11b+ 250K 105 105 105 200K 4 104 10 104 150K I 3 I 3 4 3 0 10 3 C 2 H 10 10 100K 1 S D C D S C M 2 C 10 50K 0 0 0 0 0 102 103 104 105 0 103 104 105 0 103 104 105 0 103 104 105 Auto fluor. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

A Human Population-Based Organotypic in Vitro Model for Cardiotoxicity Screening1
ALTEX preprint published July 8, 2018 doi:10.14573/altex.1805301 Research Article A human population-based organotypic in vitro model for cardiotoxicity screening1 Fabian A. Grimm1, Alexander Blanchette1, John S. House2, Kyle Ferguson1, Nan-Hung Hsieh1, Chimeddulam Dalaijamts1, Alec A. Wright1, Blake Anson5, Fred A. Wright3,4, Weihsueh A. Chiu1, Ivan Rusyn1 1Department of Veterinary Integrative Biosciences, Texas A&M University, College Station, TX, USA; 2Bioinformatics Research Center, 3Department of Biological Sciences, and 4Department of Statistics, North Carolina State University, Raleigh, NC, USA; 5Cellular Dynamics International, Madison, WI, USA Abstract Assessing inter-individual variability in responses to xenobiotics remains a substantial challenge, both in drug development with respect to pharmaceuticals and in public health with respect to environmental chemicals. Although approaches exist to characterize pharmacokinetic variability, there are no methods to routinely address pharmacodynamic variability. In this study, we aimed to demonstrate the feasibility of characterizing inter-individual variability in a human in vitro model. Specifically, we hypothesized that genetic variability across a population of iPSC- derived cardiomyocytes translates into reproducible variability in both baseline phenotypes and drug responses. We measured baseline and drug-related effects in iPSC-derived cardiomyocytes from 27 healthy donors on kinetic Ca2+ flux and high-content live cell imaging. Cells were treated in concentration-response with cardiotoxic drugs: isoproterenol (β- adrenergic receptor agonist/positive inotrope), propranolol (β-adrenergic receptor antagonist/negative inotrope), and cisapride (hERG channel inhibitor/QT prolongation). Cells from four of the 27 donors were further evaluated in terms of baseline and treatment-related gene expression. Reproducibility of phenotypic responses was evaluated across batches and time. -

ADP-Ribosylation Factor, a Small GTP-Binding Protein, Is Required for Binding of the Coatomer Protein Fl-COP to Golgi Membranes JULIE G
Proc. Natl. Acad. Sci. USA Vol. 89, pp. 6408-6412, July 1992 Biochemistry ADP-ribosylation factor, a small GTP-binding protein, is required for binding of the coatomer protein fl-COP to Golgi membranes JULIE G. DONALDSON*, DAN CASSEL*t, RICHARD A. KAHN*, AND RICHARD D. KLAUSNER* *Cell Biology and Metabolism Branch, National Institute of Child Health and Human Development, and tLaboratory of Biological Chemistry, Division of Cancer Treatment, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892 Communicated by Marc Kirschner, April 20, 1992 (receivedfor review February 11, 1992) ABSTRACT The coatomer is a cytosolic protein complex localized to the Golgi complex, although their functions have that reversibly associates with Golgi membranes and is Impli- not been defined. Distinct among these proteins is the ADP- cated in modulating Golgi membrane transport. The associa- ribosylation factor (ARF), originally identified as a cofactor tion of 13-COP, a component of coatomer, with Golgi mem- required for in vitro cholera toxin-catalyzed ADP- branes is enhanced by guanosine 5'-[v-thioltriphosphate ribosylation of the a subunit of the trimeric GTP-binding (GTP[yS]), a nonhydrolyzable analogue of GTP, and by a protein G, (G,.) (19). ARF is an abundant cytosolic protein mixture of aluminum and fluoride ions (Al/F). Here we show that reversibly associates with Golgi membranes (20, 21). that the ADP-ribosylation factor (ARF) is required for the ARF has been shown to be present on Golgi coated vesicles binding of (-COP. Thus, 13-COP contained in a coatomer generated in the presence of GTP[yS], but it is not a com- fraction that has been resolved from ARF does not bind to Golgi ponent of the cytosolic coatomer (22).