Molecular Spandrels: Tests of Adaptation at the Genetic Level
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

An Introgression Analysis of Quantitative Trait Loci That Contribute to a Morphological Difference Between Drosophila Simulans and D
Copyight 0 1997 by the Genetics Societv of America An Introgression Analysis of Quantitative Trait Loci That Contribute to a Morphological Difference Between Drosophila simulans and D. mauritiana Cathy C. Laurie, John R. True, Jianjun Liu and John M. Mercer Department of Zoology, Duke University, Durham, North Carolina 27708 Manuscript received August 12, 1996 Accepted for publication October 29, 1996 ABSTRACT Drosophila simulans and D. maum'tiana differ markedly in morphology of the posterior lobe, a male- specific genitalic structure. Bothsize and shapeof the lobe can be quantifiedby a morphometric variable, PC1, derived from principal components and Fourier analyses. The genetic architectureof the species difference in PC1 was investigated previously by composite interval mapping, which revealed largely additive inheritance, with a minimum of eight quantitative trait loci (QTL) affecting the trait. This analysis was extended by introgression of marked segments of the maun'tiana third chromosome into a simulans background by repeated backcrossing. Thetwo types of experiment are consistentin suggesting that several QTL on the third chromosome may have effects in the range of 10-15% of the parental difference andthat all or nearly all QTL have effects in the same direction. Since the parental difference is large (30.4 environmental standard deviations), effects of this magnitude can produce alternative homozygotes with little overlap in phenotype. However, these estimates may not reflect the effects of individual loci, since each interval or introgressed segment may contain multiple QTL. The consistent direction of allelic effects suggests a history of directional selection on the posterior lobe. HE genetic analysis of a quantitative trait usually ual genes that affect continuously variable traits. -

And Ford, I; Ford, '953) on the Other Hand Have Put Forward a View Intermediate Between the Extreme Ones of Darwin on the One Hand and Goldschmidt on the Other
THE EVOLUTION OF MIMICRY IN THE BUTTERFLY PAPILIO DARDANUS C. A. CLARKE and P. M. SHEPPARD Departments of Medicine and Zoology, University of Liverpool Received23.V.59 1.INTRODUCTION WHENBatesputforward the mimicry hypothesis which bears his name, Darwin (1872), although accepting it, had some difficulty in explaining the evolution of the mimetic resemblance of several distinct species to one distasteful model by a series of small changes, a require- ment of his general theory of evolution. He said "it is necessary to suppose in some cases that ancient members belonging to several distinct groups, before they had diverged to their present extent, accidentally resembled a member of another and protected group in a sufficient degree to afford some slight protection; this having given the basis for the subsequent acquisition of the most perfect resemb- lance ". Punnett (1915) realised that the difficulty is even more acute when one is dealing with a polymorphic species whose forms mimic very distantly related models. Knowing that, in those butterflies which had been investigated genetically, the forms differed by single allelomorphs he concluded that the mimicry did not evolve gradually and did not confer any advantage or disadvantage to the individual. He argued that an allelomorph arises at a single step by mutation and that therefore the mimicry also arises by chance at a single step. Goldschmidt (x) although not denying that mimicry confers some advantage to its possessors also maintained that the resemblance arises fully perfected by a single mutation of a gene distinct from that producing the colour pattern in the model, but producing a similar effect in the mimic. -

Microevolution and the Genetics of Populations Microevolution Refers to Varieties Within a Given Type
Chapter 8: Evolution Lesson 8.3: Microevolution and the Genetics of Populations Microevolution refers to varieties within a given type. Change happens within a group, but the descendant is clearly of the same type as the ancestor. This might better be called variation, or adaptation, but the changes are "horizontal" in effect, not "vertical." Such changes might be accomplished by "natural selection," in which a trait within the present variety is selected as the best for a given set of conditions, or accomplished by "artificial selection," such as when dog breeders produce a new breed of dog. Lesson Objectives ● Distinguish what is microevolution and how it affects changes in populations. ● Define gene pool, and explain how to calculate allele frequencies. ● State the Hardy-Weinberg theorem ● Identify the five forces of evolution. Vocabulary ● adaptive radiation ● gene pool ● migration ● allele frequency ● genetic drift ● mutation ● artificial selection ● Hardy-Weinberg theorem ● natural selection ● directional selection ● macroevolution ● population genetics ● disruptive selection ● microevolution ● stabilizing selection ● gene flow Introduction Darwin knew that heritable variations are needed for evolution to occur. However, he knew nothing about Mendel’s laws of genetics. Mendel’s laws were rediscovered in the early 1900s. Only then could scientists fully understand the process of evolution. Microevolution is how individual traits within a population change over time. In order for a population to change, some things must be assumed to be true. In other words, there must be some sort of process happening that causes microevolution. The five ways alleles within a population change over time are natural selection, migration (gene flow), mating, mutations, or genetic drift. -

Patterns and Power of Phenotypic Selection in Nature
Articles Patterns and Power of Phenotypic Selection in Nature JOEL G. KINGSOLVER AND DAVID W. PFENNIG Phenotypic selection occurs when individuals with certain characteristics produce more surviving offspring than individuals with other characteristics. Although selection is regarded as the chief engine of evolutionary change, scientists have only recently begun to measure its action in the wild. These studies raise numerous questions: How strong is selection, and do different types of traits experience different patterns of selection? Is selection on traits that affect mating success as strong as selection on traits that affect survival? Does selection tend to favor larger body size, and, if so, what are its consequences? We explore these questions and discuss the pitfalls and future prospects of measuring selection in natural populations. Keywords: adaptive landscape, Cope’s rule, natural selection, rapid evolution, sexual selection henotypic selection occurs when individuals with selection on traits that affect survival stronger than on those Pdifferent characteristics (i.e., different phenotypes) that affect only mating success? In this article, we explore these differ in their survival, fecundity, or mating success. The idea and other questions about the patterns and power of phe- of phenotypic selection traces back to Darwin and Wallace notypic selection in nature. (1858), and selection is widely accepted as the primary cause of adaptive evolution within natural populations.Yet Darwin What is selection, and how does it work? never attempted to measure selection in nature, and in the Selection is the nonrandom differential survival or repro- century following the publication of On the Origin of Species duction of phenotypically different individuals. -

The Spandrels of Self-Deception: Prospects for a Biological Theory of a Mental Phenomenon
The Spandrels of Self-Deception: Prospects for a Biological Theory of a Mental Phenomenon D. S. Neil Van Leeuwen Self-deception is so undeniable a fact of human life that if anyone tried to deny its existence, the proper response would be to accuse this person of it. —Allen Wood1 1. Introduction to the Problems Even prior to a characterization of self-deception, it’s easy to see that it’s widespread. This is puzzling, because self-deception seems paradoxical. Here are three apparent problems. First, self-deception seems to involve a conceptual contradiction; in order to deceive, one must believe the contrary of the deception one is perpetrating, but if one believes the contrary, it seems impossible for that very self to believe the deception.2 Second, a view of the mind has become widely accepted that makes rationality constitutive and exhaustive of the mental; this is the interpretive view of the mental put forth by Donald Davidson and followers.3 According to this view, the idea of attributing irrational beliefs to agents makes little sense, for we cannot make sense of what someone’s beliefs are unless they are rational. But self-deception is highly irrational.4 Third, a brief survey of other cognitive mechanisms (such as vision) reveals that most are well suited for giving us reliable information about ourselves and our environment. Self- deception, however, undermines knowledge of ourselves and the world. If having good information brings fitness benefits to organisms, how is it that self-deception was not weeded out by natural selection? Otherwise put, why does self-deception exist? (For convenience, I shall refer to these as puzzles 1, 2, and 3.) Puzzles such as these make self-deception an intriguing explanatory target. -

Arise by Chance As the Result of Mutation. They Therefore Suggest
THE EVOLUTION OF DOMINANCE UNDER DISRUPTIVE SELECTION C. A. CLARKE and P. Ni. SHEPPARD Department of Medicine and Department of Zoology, University of Liverpool Received6.iii.59 1.INTRODUCTION INa paper on the effects of disruptive selection, Mather (1955) pointed out that if there are two optimum values for a character and all others are less advantageous or disadvantageous there will be disruptive selection which can lead to the evolution of a polymorphism. Sheppard (1958) argued that where such selection is effective and the change from one optimum value to the other is switched by a single pair of allelomorphs there will be three genotypes but only two advantageous phenotypes. Consequently if dominance were absent initially it would be evolved as a result of the disruptive selection, the heterozygote and one of the homozygotes both coming to resemble one of the two optimum phenotypes (see Ford, 1955, on Tripharna comes). Thoday (1959) has shown by means of an artificial selection experiment that, even when a character is, at the beginning, controlled polygenically (sternopleural chaeta-number in Drosophila) and there is 50 per cent. gene exchange between the "high" and "low" selected sub-popu- lations, a polymorphism can evolve. The most fully understood examples of disruptive selection (other than sex) are provided by instances of Batesian Mimicry, where there are a number of distinct warningly coloured species, acting as models, which are mimicked by the polymorphic forms of a single more edible species. Fisher and Ford (see Ford, 1953) have argued that a suffi- ciently good resemblance between mimic and model is not likely to arise by chance as the result of mutation. -
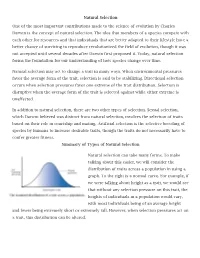
Natural Selection One of the Most Important Contributions Made to The
Natural Selection One of the most important contributions made to the science of evolution by Charles Darwin is the concept of natural selection. The idea that members of a species compete with each other for resources and that individuals that are better adapted to their lifestyle have a better chance of surviving to reproduce revolutionized the field of evolution, though it was not accepted until several decades after Darwin first proposed it. Today, natural selection forms the foundation for our understanding of how species change over time. Natural selection may act to change a trait in many ways. When environmental pressures favor the average form of the trait, selection is said to be stabilizing. Directional selection occurs when selection pressures favor one extreme of the trait distribution. Selection is disruptive when the average form of the trait is selected against while either extreme is unaffected. In addition to natural selection, there are two other types of selection. Sexual selection, which Darwin believed was distinct from natural selection, involves the selection of traits based on their role in courtship and mating. Artificial selection is the selective breeding of species by humans to increase desirable traits, though the traits do not necessarily have to confer greater fitness. Summary of Types of Natural Selection Natural selection can take many forms. To make talking about this easier, we will consider the distribution of traits across a population in using a graph. To the right is a normal curve. For example, if we were talking about height as a trait, we would see that without any selection pressure on this trait, the heights of individuals in a population would vary, with most individuals being of an average height and fewer being extremely short or extremely tall. -

The Spandrels of San Marco and the Panglossian Paradigm: a Critique of the Adaptationist Programme
The Spandrels of San Marco and the Panglossian Paradigm: A Critique of the Adaptationist Programme. (Excerpted1 by W. M. Schaffer) Stephen Jay Gould and Richard C. Lewontin. 1979. Proc. Roy. Soc. London, Ser. B, 505: 581-598. An adaptationist programme has dominated evolutionary thought in England and the United States during the past forty years. It is based on faith in the power of natural selection as an optimizing agent. It proceeds by breaking an organism into unitary "traits" and proposing an adaptive story for each considered separately. Trade-offs among competing selective demands exert the only brake upon perfection; non-optimality is thereby rendered as a result of adaptation as well. We criticize this approach and attempt to reassert a competing notion (long popular in continental Europe) that organisms must be analyzed as integrated wholes, with Baupläne[2] so constrained by phyletic heritage, pathways of development, and general architecture that the constraints them- selves become more interesting and more important in delimiting pathways of change than the selective force that may mediate change when it occurs. We fault the adaptationist programme for its failure to distinguish current utility from reasons for origin (male tyrannosaurs may have used their diminutive front legs to titillate female partners, but this will not explain why they got so small); for its unwillingness to consider alternatives to adaptive stories; for its reliance upon plau- sibility alone as a criterion for accepting speculative tales; and for its failure to consider adequately such competing themes as random fixation of alleles, production of non-adaptive structures by developmental correlation with selected features … , the separability of adaptation and selection, multiple adaptive peaks, and current utility as an epiphenomenon of nonadaptive structures. -

Supplementary Information for Vyssotski, 2013
Supplementary Information for Vyssotski, 2013 www.evolocus.com/evolocus/v1/evolocus-01-013-s.pdf Evolocus Supplementary Information for Transgenerational epigenetic compensation and sex u al d imorph ism 1,2,3 Dmitri L. Vyssotski Published 17 May 2013, Evolocus 1, 13-18 (2013): http://www.evolocus.com/evolocus/v1/evolocus-01-013.pdf In natu ral popu lation transgenerational epigenetic compensation is by unknown mechanisms, the mechanisms those were difficult to generated in h omoz y gou s mu tants, mainly in males. It d oes not imagine, and this possibility was abandoned for over 47 years. immed iately affect th e ph enoty pe of h omoz y gou s mu tants, w h ere The higher variability in males can be a result of a not so good it w as generated . Transgenerational epigenetic compensation is canalization of their ontogenesis, and the canalization of localiz ed not in th e same locu s as original mu tation and it is ontogenesis is modulated by sex hormones. Thus, we can d ominant, b ecau se it affects th e ph enoty pe ev en if it is receiv ed imagine that the canalization of ontogenesis in males is from one parent. In fu rth er generations, starting from F 2, it is more artificially decreased in comparison with females by means of ex pressed in females th an in males: it increases fitness of hormonal differences. h omozy gou s mu tant females, d ecreases fitness of w ild ty pe The shift of the female distribution to the right was not females and h as w eak effect on h eterozy gou s females. -

Stabilizing Selection Shapes Variation in Phenotypic Plasticity
bioRxiv preprint doi: https://doi.org/10.1101/2021.07.29.454146; this version posted July 29, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Stabilizing selection shapes variation in phenotypic plasticity 2 3 4 5 Dörthe Becker1,2,*, 6 Karen Barnard-Kubow1,3, 7 Robert Porter1, 8 Austin Edwards1,4, 9 Erin Voss1,5, 10 Andrew P. Beckerman2, 11 Alan O. Bergland1,* 12 13 14 1Department of Biology; University of Virginia, Charlottesville/VA, USA 15 2Department of Animal and Plant Sciences; University of Sheffield, Sheffield, UK 16 3Department of Biology, James Madison University, Harrisonburg/VA, USA 17 4Biological Imaging Development CoLab, University of California San Francisco, 18 San Francisco/CA, USA 19 5Department of Integrative Biology, University of California Berkeley, 20 Berkeley/CA 21 22 * to whom correspondence should be addressed: 23 DB ([email protected]), AOB ([email protected]) 24 25 26 27 Keywords: stabilizing selection; phenotypic plasticity; heritability; Daphnia; 28 defense morphologies 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.07.29.454146; this version posted July 29, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 29 Abstract 30 The adaptive nature of phenotypic plasticity is widely documented in natural 31 populations. However, little is known about the evolutionary forces that shape 32 genetic variation in plasticity within populations. Here we empirically address this 33 issue by testing the hypothesis that stabilizing selection shapes genetic variation 34 in the anti-predator developmental plasticity of Daphnia pulex. -

Exaptation, Adaptation, and Evolutionary Psychology
Exaptation, Adaptation, and Evolutionary Psychology Armin Schulz Department of Philosophy, Logic, and Scientific Method London School of Economics and Political Science Houghton St London WC2A 2AE UK [email protected] (0044) 753-105-3158 Abstract One of the most well known methodological criticisms of evolutionary psychology is Gould’s claim that the program pays too much attention to adaptations, and not enough to exaptations. Almost as well known is the standard rebuttal of that criticism: namely, that the study of exaptations in fact depends on the study of adaptations. However, as I try to show in this paper, it is premature to think that this is where this debate ends. First, the notion of exaptation that is commonly used in this debate is different from the one that Gould and Vrba originally defined. Noting this is particularly important, since, second, the standard reply to Gould’s criticism only works if the criticism is framed in terms of the former notion of exaptation, and not the latter. However, third, this ultimately does not change the outcome of the debate much, as evolutionary psychologists can respond to the revamped criticism of their program by claiming that the original notion of exaptation is theoretically and empirically uninteresting. By discussing these issues further, I also seek to determine, more generally, which ways of approaching the adaptationism debate in evolutionary biology are useful, and which not. Exaptation, Adaptation, and Evolutionary Psychology Exaptation, Adaptation, and Evolutionary Psychology I. Introduction From a methodological point of view, one of the most well known accusations of evolutionary psychology – the research program emphasising the importance of appealing to evolutionary considerations in the study of the mind – is the claim that it is overly “adaptationist” (for versions of this accusation, see e.g. -

Exaptation in Human Evolution: How to Test Adaptive Vs Exaptive Evolutionary Hypotheses
doi 10.4436/jass.89015 JASs Invited Reviews Journal of Anthropological Sciences Vol. 89 (2011), pp. 9-23 Exaptation in human evolution: how to test adaptive vs exaptive evolutionary hypotheses Telmo Pievani & Emanuele Serrelli University of Milan Bicocca, Piazza dell’Ateneo Nuovo, n. 1 - 20126 Milan, Italy e-mail: [email protected]; [email protected] Summary - Palaeontologists, Stephen J. Gould and Elisabeth Vrba, introduced the term “ex-aptation” with the aim of improving and enlarging the scientific language available to researchers studying the evolution of any useful character, instead of calling it an “adaptation” by default, coming up with what Gould named an “extended taxonomy of fitness” . With the extension to functional co-optations from non-adaptive structures (“spandrels”), the notion of exaptation expanded and revised the neo-Darwinian concept of “pre- adaptation” (which was misleading, for Gould and Vrba, suggesting foreordination). Exaptation is neither a “saltationist” nor an “anti-Darwinian” concept and, since 1982, has been adopted by many researchers in evolutionary and molecular biology, and particularly in human evolution. Exaptation has also been contested. Objections include the “non-operationality objection”.We analyze the possible operationalization of this concept in two recent studies, and identify six directions of empirical research, which are necessary to test “adaptive vs. exaptive” evolutionary hypotheses. We then comment on a comprehensive survey of literature (available online), and on the basis of this we make a quantitative and qualitative evaluation of the adoption of the term among scientists who study human evolution. We discuss the epistemic conditions that may have influenced the adoption and appropriate use of exaptation, and comment on the benefits of an “extended taxonomy of fitness” in present and future studies concerning human evolution.